STATS 220
Tidy data🧹
1 / 36
2 / 36
2 / 36
image sources: Upsplash
2 / 36
- each piece is a scalar, a vector, a tibble, a list
- each piece is vector functions or table functions
- bc they are standard and consistent blocks,
- ensemble them to a simple mario, or complex nintendo
- so far you worked with several datasets now,
- hopefully you find a pattern is using these dplyr verbs in different ways to solve various problems.
What you think about data science 🤩
image credit: Hilary Parker
3 / 36
- 80% on modelling
What data science is in reality 😭
image credit: Hilary Parker
4 / 36
- 80% on cleaning
- it's invaluable to know the data pipeline: how data collected and organised
- covid19, different def for each country in the early stage
tidy data clean data
The movies data is tidy but not clean.
#> # A tibble: 5 x 16#> Release_Date US_DVD_Sales Title US_Gross Worldwide_Gross Production_Budg…#> <chr> <int> <chr> <int> <dbl> <int>#> 1 9-Mar-94 NA Four Wedd… 52700832 242895809 4500000#> 2 18-Oct-06 NA 51 Birch … 84689 84689 350000#> 3 1963-01-01 NA 55 Days a… 10000000 10000000 17000000#> 4 <NA> NA Drei 0 0 7200000#> 5 16-Jan-98 NA The Dress 16556 16556 2650000#> # … with 10 more variables: MPAA_Rating <chr>, Running_Time_min <int>,#> # Distributor <chr>, Source <chr>, Major_Genre <chr>, Creative_Type <chr>,#> # Director <chr>, Rotten_Tomatoes_Rating <int>, IMDB_Rating <dbl>,#> # IMDB_Votes <int>5 / 36
- all data you've seen so far are tidy, but tidy doesn't mean clean.
- how I would clean this data:
- either google Drei release year, and corresponding GDP
- or replace GDP with NA
- Gross are suspicious
tidy data clean data
They are tidy and clean in their own way.
time_use
#> # A tibble: 28 x 3#> country category minutes#> <chr> <chr> <dbl>#> 1 New Zeala… Paid work 241 #> 2 USA Paid work 251. #> 3 New Zeala… Education 29 #> 4 USA Education 31.4#> 5 New Zeala… Care for household… 30 #> 6 USA Care for household… 30.6#> # … with 22 more rowscountrycode
#> # A tibble: 2 x 2#> country country_name #> <chr> <chr> #> 1 NZL New Zealand #> 2 USA United States6 / 36
- week 4, when we try to join them, we see how country names are encoded differently.
- Data cleaning is laborious, and highly dependent on the data itself.
- No golden standards or principles to guide how to clean data.
- reason about the cleaning steps (intuition, common sense, business context)
- principles to get tidy data.
Tidy data describes a standard way of storing data in a consistent format.
7 / 36
- tidy data is about data format, instead of caring the data values.
Your turn
Tuberculosis data from WHO (data/tb.csv):
- Is
tbtidy data? - What are the data observations? What are the data variables?
tb#> # A tibble: 5,769 x 22#> iso2 year m_04 m_514 m_014 m_1524 m_2534 m_3544 m_4554 m_5564 m_65 m_u#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>#> 1 AD 1989 NA NA NA NA NA NA NA NA NA NA#> 2 AD 1990 NA NA NA NA NA NA NA NA NA NA#> 3 AD 1991 NA NA NA NA NA NA NA NA NA NA#> 4 AD 1992 NA NA NA NA NA NA NA NA NA NA#> 5 AD 1993 NA NA NA NA NA NA NA NA NA NA#> 6 AD 1994 NA NA NA NA NA NA NA NA NA NA#> # … with 5,763 more rows, and 10 more variables: f_04 <dbl>, f_514 <dbl>,#> # f_014 <dbl>, f_1524 <dbl>, f_2534 <dbl>, f_3544 <dbl>, f_4554 <dbl>,#> # f_5564 <dbl>, f_65 <dbl>, f_u <dbl>01:00
8 / 36
- Can you easily plot a dataset like this using ggplot2? by sex and age
- come back later
one data, many representations

image credit: Garrick Aden-Buie
9 / 36
- which rep does
tbresemble? - which rep is easier to work with? depends on the question, wide -> suitable for summary table
10 / 36
- In database, this is schema.
- Tidy data principles are a rephrase of third norm in a database schema design. https://en.wikipedia.org/wiki/Third_normal_form, to data scientists.
- tidy data is for human consumption.
- Tabular data is column-oriented format
11 / 36
Making friends with tidy data
- one set of consistent tools for different datasets
- easier for automation and iteration
reference: Illustrations from the Openscapes blog Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst
12 / 36
- To work with messy data, every time you need to switch to different gears
- learn new tools that just works for that specific dataset
- It's much more pleasant to work with tidy data, help you to build a good taste of data analysis
- spent less flighting with different tools, focus more on data analysis bc one set of consistent tools
- the {tidyverse} philosophy to work with the tidy data structures
- build automatic workflow for analysis, feed different data sets.
- missing verbs not exactly a pair
- Messy
#> # A tibble: 5,769 x 22#> iso2 year m_04 m_514 m_014#> <chr> <dbl> <dbl> <dbl> <dbl>#> 1 AD 1989 NA NA NA#> 2 AD 1990 NA NA NA#> 3 AD 1991 NA NA NA#> 4 AD 1992 NA NA NA#> 5 AD 1993 NA NA NA#> 6 AD 1994 NA NA NA#> # … with 5,763 more rows, and 17#> # more variables: m_1524 <dbl>,#> # m_2534 <dbl>, m_3544 <dbl>,#> # m_4554 <dbl>, m_5564 <dbl>,#> # m_65 <dbl>, m_u <dbl>,#> # f_04 <dbl>, f_514 <dbl>,#> # f_014 <dbl>, f_1524 <dbl>,#> # f_2534 <dbl>, f_3544 <dbl>,#> # f_4554 <dbl>, f_5564 <dbl>,#> # f_65 <dbl>, f_u <dbl>- Tidy
#> # A tibble: 115,380 x 5#> iso2 year sex age cases#> <chr> <dbl> <chr> <chr> <dbl>#> 1 AD 1989 m 04 0#> 2 AD 1989 m 514 0#> 3 AD 1989 m 014 0#> 4 AD 1989 m 1524 0#> 5 AD 1989 m 2534 0#> 6 AD 1989 m 3544 0#> # … with 115,374 more rows14 / 36
- the
tbdata is messy data, rows and columns - obs and variables
- working with tidy data, we know obs: the number of cases by sex and age groups over years at country level
- messy in a way that column names should be values, stored in cells.
- what steps needed to get to this tidy format.
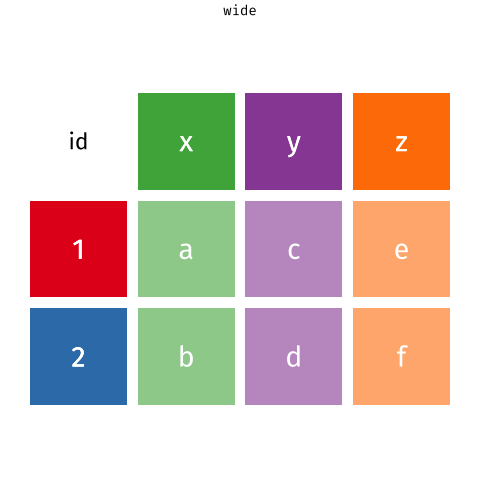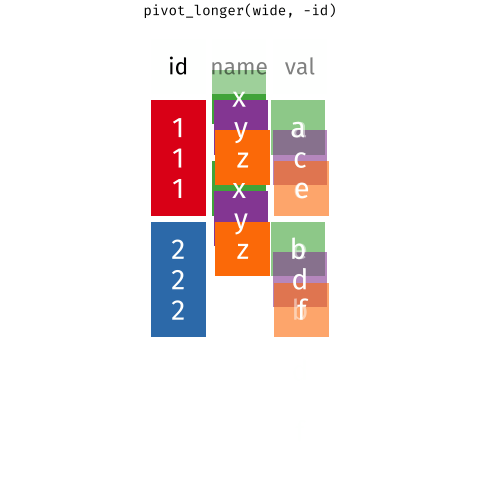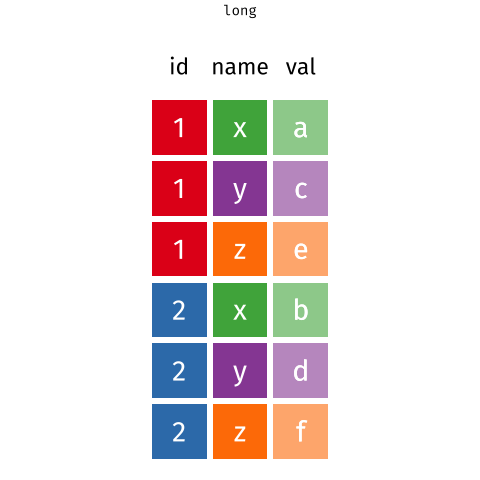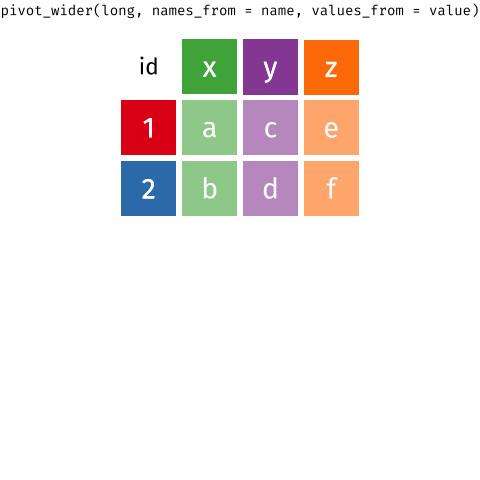
pivot_longer()a wider-format data
library(tidyverse) # library(tidyr)tb %>% pivot_longer( cols = m_04:f_u, # cols in the data for pivoting names_to = "sex_age", # new col contains old headers values_to = "cases") # new col contains old values#> # A tibble: 115,380 x 4#> iso2 year sex_age cases#> <chr> <dbl> <chr> <dbl>#> 1 AD 1989 m_04 NA#> 2 AD 1989 m_514 NA#> 3 AD 1989 m_014 NA#> 4 AD 1989 m_1524 NA#> 5 AD 1989 m_2534 NA#> 6 AD 1989 m_3544 NA#> # … with 115,374 more rows15 / 36
- messy: store two variables into one column
seperate()a character column into multiple columns
tb %>% pivot_longer(cols = m_04:f_u, names_to = "sex_age", values_to = "cases") %>% separate(sex_age, into = c("sex", "age"), sep = "_")#> # A tibble: 115,380 x 5#> iso2 year sex age cases#> <chr> <dbl> <chr> <chr> <dbl>#> 1 AD 1989 m 04 NA#> 2 AD 1989 m 514 NA#> 3 AD 1989 m 014 NA#> 4 AD 1989 m 1524 NA#> 5 AD 1989 m 2534 NA#> 6 AD 1989 m 3544 NA#> # … with 115,374 more rows17 / 36
fill()inNAwith previous ("down") or next ("up") value
tb_tidy <- tb %>% pivot_longer(cols = m_04:f_u, names_to = "sex_age", values_to = "cases") %>% separate(sex_age, into = c("sex", "age"), sep = "_") %>% group_by(iso2) %>% fill(cases, .direction = "updown") %>% ungroup()#> # A tibble: 115,380 x 5#> iso2 year sex age cases#> <chr> <dbl> <chr> <chr> <dbl>#> 1 AD 1989 m 04 0#> 2 AD 1989 m 514 0#> 3 AD 1989 m 014 0#> 4 AD 1989 m 1524 0#> 5 AD 1989 m 2534 0#> 6 AD 1989 m 3544 0#> # … with 115,374 more rows18 / 36
- how to impute missing data is dependent on data context. For cross-sectional, average and build a regression model to predict
- fill by direction
- demo on
direction
nest()multiple columns into a list-column
tb_tidy %>% nest(data = -iso2)#> # A tibble: 213 x 2#> iso2 data #> <chr> <list> #> 1 AD <tibble[,4] [380 × 4]>#> 2 AE <tibble[,4] [520 × 4]>#> 3 AF <tibble[,4] [480 × 4]>#> 4 AG <tibble[,4] [540 × 4]>#> 5 AI <tibble[,4] [440 × 4]>#> 6 AL <tibble[,4] [540 × 4]>#> # … with 207 more rows19 / 36
- tibble is a list, recursive structure
- we can put a list into a tibble
- complex objs in tibble
Building many models*
tb_fit <- tb_tidy %>% nest(data = -iso2) %>% mutate( # map() for week 11 model = map(data, ~ lm(cases ~ year + sex + age, data = .)), r2 = map_dbl(model, ~ broom::glance(.)$r.squared) )#> # A tibble: 213 x 4#> iso2 data model r2#> <chr> <list> <list> <dbl>#> 1 AD <tibble[,4] [380 × 4]> <lm> 0.165#> 2 AE <tibble[,4] [520 × 4]> <lm> 0.133#> 3 AF <tibble[,4] [480 × 4]> <lm> 0.487#> 4 AG <tibble[,4] [540 × 4]> <lm> 0.220#> 5 AI <tibble[,4] [440 × 4]> <lm> 0.515#> 6 AL <tibble[,4] [540 × 4]> <lm> 0.478#> # … with 207 more rows20 / 36
- r2 for each country
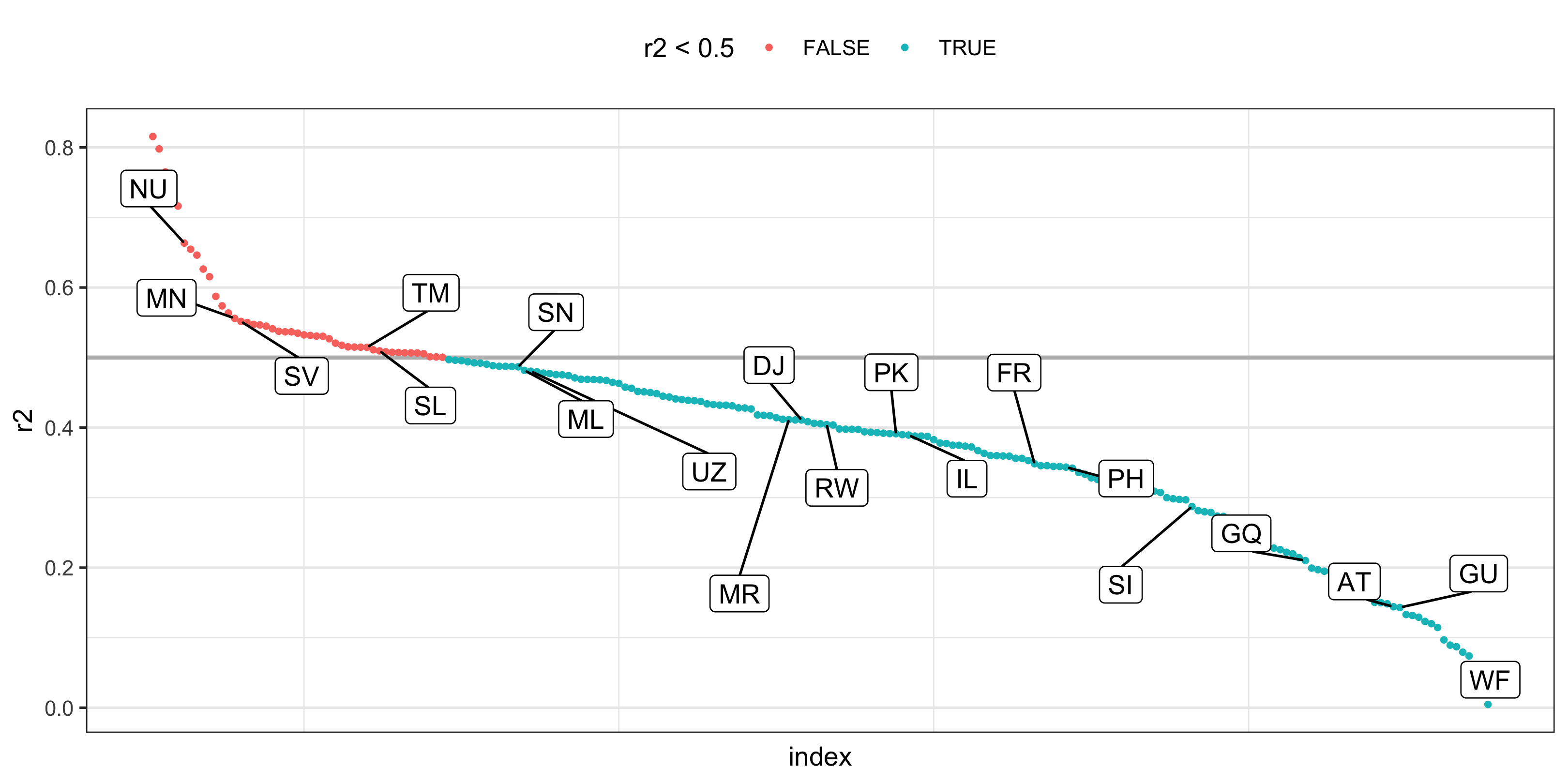
set.seed(220)tb_fit_pre <- tb_fit %>% arrange(-r2) %>% mutate(iso2 = fct_inorder(iso2), index = row_number())hl_tb <- tb_fit_pre %>% slice_sample(n = 20)tb_fit_pre %>% ggplot(aes(x = index, y = r2)) + geom_hline(yintercept = 0.5, size = 0.8, colour = "grey") + geom_point(aes(colour = r2 < 0.5), size = 0.8) + ggrepel::geom_label_repel(aes(label = iso2), data = hl_tb, box.padding = 1) + theme_bw() + theme( axis.text.x = element_blank(), axis.ticks.x = element_blank(), panel.grid.major.x = element_blank(), legend.position = "top" )21 / 36
- direct labeling instead of > 200 country labels underneath
Your turn
Auckland weather data from GHCND (data/ghcnd/ghcnd-akl.csv):
- Is
aklweathertidy data? - What are the data observations? What are the data variables?
aklweather#> # A tibble: 2,974 x 3#> date datatype value#> <date> <chr> <dbl>#> 1 2019-01-01 PRCP 0#> 2 2019-01-01 TAVG 206#> 3 2019-01-01 TMAX 232#> 4 2019-01-01 TMIN 188#> 5 2019-01-02 PRCP 5#> 6 2019-01-02 TAVG 207#> # … with 2,968 more rows01:00
22 / 36
- The
valuecolumn contains multiple types of measurements.
- Messy
#> # A tibble: 2,974 x 3#> date datatype value#> <date> <chr> <dbl>#> 1 2019-01-01 PRCP 0#> 2 2019-01-01 TAVG 206#> 3 2019-01-01 TMAX 232#> 4 2019-01-01 TMIN 188#> 5 2019-01-02 PRCP 5#> 6 2019-01-02 TAVG 207#> # … with 2,968 more rowsaklweather <- read_csv2( "data/ghcnd/ghcnd-akl.csv", col_types = cols_only( date = "D", datatype = "c", value = "d"))aklweather- Tidy
#> # A tibble: 816 x 5#> date PRCP TAVG TMAX TMIN#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2019-01-01 0 206 232 188#> 2 2019-01-02 5 207 230 183#> 3 2019-01-03 0 211 241 184#> 4 2019-01-04 0 192 229 NA#> 5 2019-01-05 0 200 233 150#> 6 2019-01-06 0 213 237 169#> # … with 810 more rows23 / 36
- demo on
read_csv2() - daily weather information in Auckland
- temp values unbelievably large?
- calibrate the measurements
aklweather %>% mutate(value = value / 10)#> # A tibble: 2,974 x 3#> date datatype value#> <date> <chr> <dbl>#> 1 2019-01-01 PRCP 0 #> 2 2019-01-01 TAVG 20.6#> 3 2019-01-01 TMAX 23.2#> 4 2019-01-01 TMIN 18.8#> 5 2019-01-02 PRCP 0.5#> 6 2019-01-02 TAVG 20.7#> # … with 2,968 more rows24 / 36
pivot_wider()a longer-format data
aklweather %>% mutate(value = value / 10) %>% pivot_wider( names_from = datatype, # new headers from old `datatype` val values_from = value) # new col contains old `value`#> # A tibble: 816 x 5#> date PRCP TAVG TMAX TMIN#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2019-01-01 0 20.6 23.2 18.8#> 2 2019-01-02 0.5 20.7 23 18.3#> 3 2019-01-03 0 21.1 24.1 18.4#> 4 2019-01-04 0 19.2 22.9 NA #> 5 2019-01-05 0 20 23.3 15 #> 6 2019-01-06 0 21.3 23.7 16.9#> # … with 810 more rows25 / 36
rename_with()renames columns using a function
aklweather_tidy <- aklweather %>% mutate(value = value / 10) %>% pivot_wider( names_from = datatype, values_from = value) %>% rename_with(tolower)aklweather_tidy#> # A tibble: 816 x 5#> date prcp tavg tmax tmin#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2019-01-01 0 20.6 23.2 18.8#> 2 2019-01-02 0.5 20.7 23 18.3#> 3 2019-01-03 0 21.1 24.1 18.4#> 4 2019-01-04 0 19.2 22.9 NA #> 5 2019-01-05 0 20 23.3 15 #> 6 2019-01-06 0 21.3 23.7 16.9#> # … with 810 more rows27 / 36
tedious to type
complete()data with missing combinations of data
library(lubridate)aklweather_tidy %>% complete(date = full_seq( ymd(c("2019-01-01", "2021-04-01")), 1))#> # A tibble: 822 x 5#> date prcp tavg tmax tmin#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2019-01-01 0 20.6 23.2 18.8#> 2 2019-01-02 0.5 20.7 23 18.3#> 3 2019-01-03 0 21.1 24.1 18.4#> 4 2019-01-04 0 19.2 22.9 NA #> 5 2019-01-05 0 20 23.3 15 #> 6 2019-01-06 0 21.3 23.7 16.9#> # … with 816 more rows28 / 36
- implicit missing records before
aklweather_tidy %>% complete(date = full_seq( ymd(c("2019-01-01", "2021-04-01")), 1)) %>% anti_join(aklweather_tidy)#> # A tibble: 6 x 5#> date prcp tavg tmax tmin#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2019-02-16 NA NA NA NA#> 2 2019-02-17 NA NA NA NA#> 3 2019-02-18 NA NA NA NA#> 4 2021-02-11 NA NA NA NA#> 5 2021-02-12 NA NA NA NA#> 6 2021-02-20 NA NA NA NA29 / 36
implicit bf, explicit after complete()
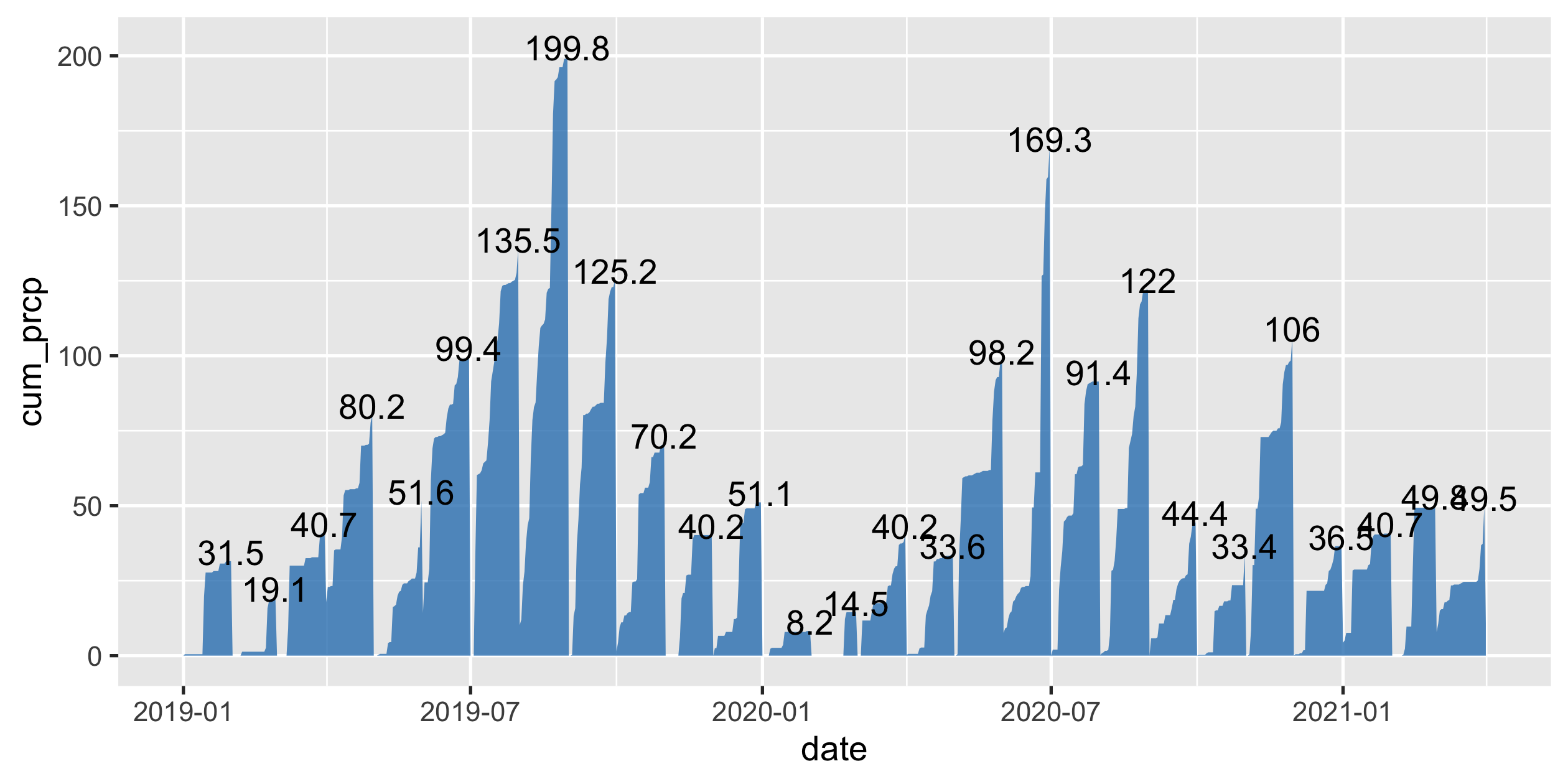
akl_prcp <- aklweather_tidy %>% complete( date = full_seq(ymd(c("2019-01-01", "2021-04-01")), 1), fill = list(prcp = 0) ) %>% group_by(yearmonth = floor_date(date, "1 month")) %>% mutate(cum_prcp = cumsum(prcp)) %>% ungroup()akl_prcp#> # A tibble: 822 x 7#> date prcp tavg tmax tmin yearmonth cum_prcp#> <date> <dbl> <dbl> <dbl> <dbl> <date> <dbl>#> 1 2019-01-01 0 20.6 23.2 18.8 2019-01-01 0 #> 2 2019-01-02 0.5 20.7 23 18.3 2019-01-01 0.5#> 3 2019-01-03 0 21.1 24.1 18.4 2019-01-01 0.5#> 4 2019-01-04 0 19.2 22.9 NA 2019-01-01 0.5#> 5 2019-01-05 0 20 23.3 15 2019-01-01 0.5#> 6 2019-01-06 0 21.3 23.7 16.9 2019-01-01 0.5#> # … with 816 more rows30 / 36
00:30
Your turn
- Why do I fill
0to missingprcp, instead of leavingNAas is?- Why do I calculate monthly cumulative precipitations?
aklweather_tidy %>% complete(date = full_seq( ymd(c("2019-01-01", "2021-04-01")), 1)) %>% filter(is.na(prcp))#> # A tibble: 13 x 5#> date prcp tavg tmax tmin#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2019-02-16 NA NA NA NA #> 2 2019-02-17 NA NA NA NA #> 3 2019-02-18 NA NA NA NA #> 4 2019-07-01 NA 13.4 18.2 NA #> 5 2019-07-02 NA 15.4 17 11.6#> 6 2019-07-03 NA 16.8 18.2 13.6#> 7 2019-11-01 NA 14.4 17 12.8#> 8 2019-11-29 NA 19.7 25.1 NA #> 9 2020-04-07 NA 16.9 22.3 NA #> 10 2020-10-20 NA 17 NA NA #> 11 2021-02-11 NA NA NA NA #> 12 2021-02-12 NA NA NA NA #> 13 2021-02-20 NA NA NA NA31 / 36
Now you need to think why I make this decision not that decision?
Area plot for monthly cumulative rainfall
32 / 36
- felt super try in 2020 summer
- Rainfall in last summer lower than the year before
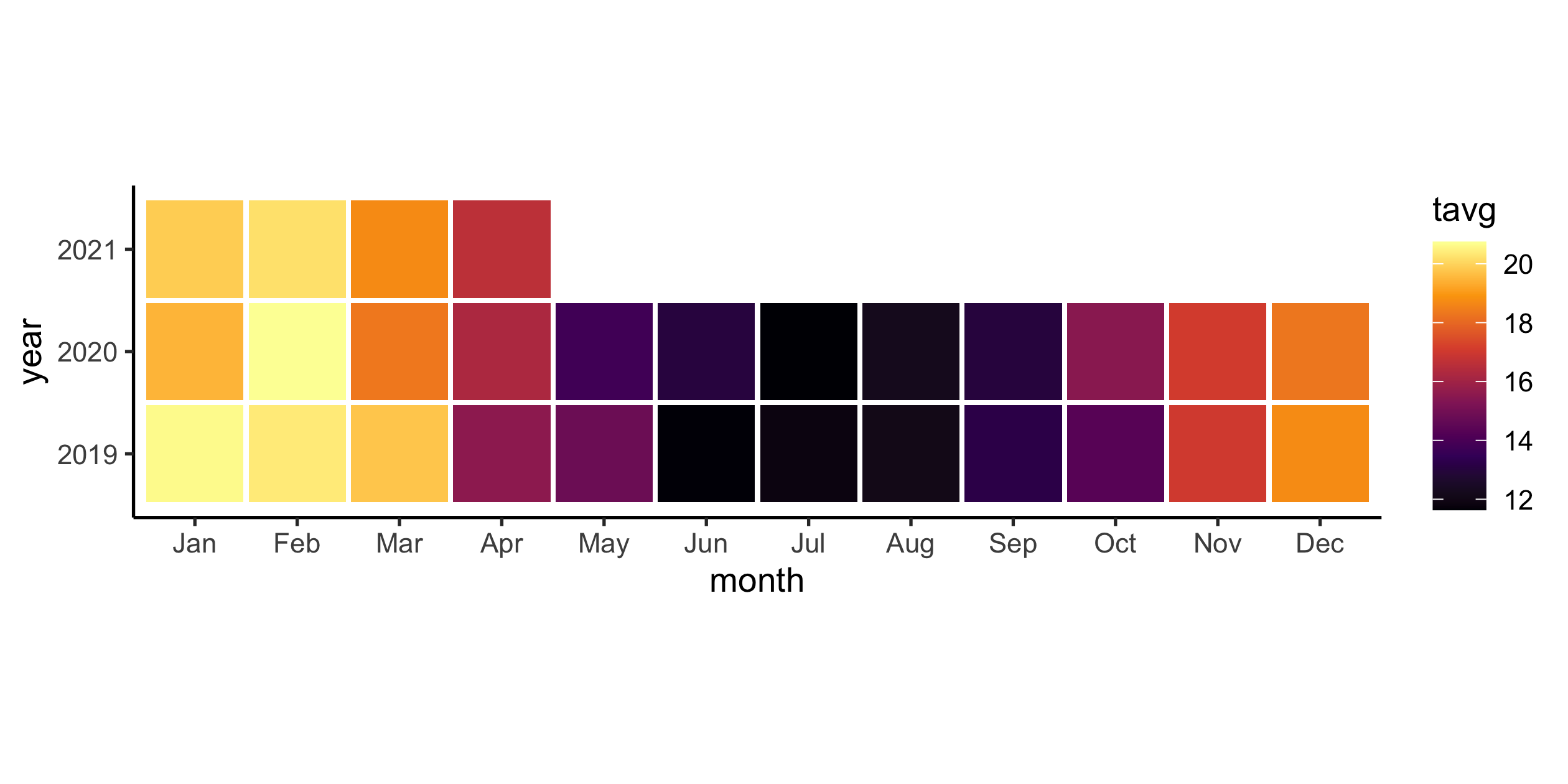
akl_monthly_temp <- aklweather_tidy %>% group_by(yearmonth = floor_date(date, "1 month")) %>% summarise( tavg = mean(tavg, na.rm = TRUE), tmax = mean(tmax, na.rm = TRUE), tmin = mean(tmin, na.rm = TRUE) )akl_monthly_temp#> # A tibble: 28 x 4#> yearmonth tavg tmax tmin#> <date> <dbl> <dbl> <dbl>#> 1 2019-01-01 20.6 24.4 17.3 #> 2 2019-02-01 20.4 25.2 16.1 #> 3 2019-03-01 19.7 24.5 15.6 #> 4 2019-04-01 15.6 20.0 10.5 #> 5 2019-05-01 14.8 18.9 10.2 #> 6 2019-06-01 11.7 15.4 7.62#> # … with 22 more rows33 / 36
Why ribbon is missing in the end?
Heatmap for monthly average temperatures
35 / 36