STATS 220
Don’t repeat yourself 🐈
Keep calm, and import CSVs one by one
library(tidyverse) # library(purrr)covid_0301 <- read_csv("https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/03-01-2020.csv")covid_0302 <- read_csv("https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/03-02-2020.csv")covid_0303 <- read_csv("https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/03-03-2020.csv")(covid_03 <- bind_rows(covid_0301, covid_0302, covid_0303))#> # A tibble: 420 x 8#> `Province/State` `Country/Region` `Last Update` #> <chr> <chr> <dttm> #> 1 Hubei Mainland China 2020-03-01 10:13:19#> 2 <NA> South Korea 2020-03-01 23:43:03#> 3 <NA> Italy 2020-03-01 23:23:02#> 4 Guangdong Mainland China 2020-03-01 14:13:18#> 5 Henan Mainland China 2020-03-01 14:13:18#> 6 Zhejiang Mainland China 2020-03-01 10:13:33#> # … with 414 more rows, and 5 more variables:#> # Confirmed <dbl>, Deaths <dbl>, Recovered <dbl>,#> # Latitude <dbl>, Longitude <dbl>Data is changing
covid_1231 <- read_csv("https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/12-31-2020.csv")covid_1231#> # A tibble: 3,980 x 14#> FIPS Admin2 Province_State Country_Region #> <dbl> <chr> <chr> <chr> #> 1 NA <NA> <NA> Afghanistan #> 2 NA <NA> <NA> Albania #> 3 NA <NA> <NA> Algeria #> 4 NA <NA> <NA> Andorra #> 5 NA <NA> <NA> Angola #> 6 NA <NA> <NA> Antigua and Barbuda#> # … with 3,974 more rows, and 10 more variables:#> # Last_Update <dttm>, Lat <dbl>, Long_ <dbl>,#> # Confirmed <dbl>, Deaths <dbl>, Recovered <dbl>,#> # Active <dbl>, Combined_Key <chr>, Incident_Rate <dbl>,#> # Case_Fatality_Ratio <dbl>What are the repeated patterns?
- everyday different URLs
- read each csv file into R
- non-standardised column names
DRY: automate the common tasks above
1 generate a sequence of urls
make_url <- function(date) { date_chr <- format(date, "%m-%d-%Y") setNames(glue::glue("https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/{date_chr}.csv"), date_chr)}3 key steps to creating a custom function:
- pick a name for the function, e.g.
make_url. - list the inputs, or arguments, to the function inside
function, e.g.date. - place the code you have developed in body of the function, a
{block that immediately followsfunction(...).
Test make_url(): does it return what we expected?
library(lubridate)dates <- seq(mdy("03-01-2020"), mdy("12-31-2020"), by = 1)urls <- make_url(dates)head(urls, 2)#> https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/03-01-2020.csv#> https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/03-02-2020.csvtail(urls, 2)#> https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/12-30-2020.csv#> https://github.com/CSSEGISandData/COVID-19/raw/master/csse_covid_19_data/csse_covid_19_daily_reports/12-31-2020.csv2 & 3 read data and standardise column names
read_covid19 <- function(url) { data <- read_csv(url, col_types = cols(Last_Update = "_", `Last Update` = "_")) rename_with(data, janitor::make_clean_names)}read_covid19(urls[1])#> # A tibble: 126 x 7#> province_state country_region confirmed deaths recovered#> <chr> <chr> <dbl> <dbl> <dbl>#> 1 Hubei Mainland China 66907 2761 31536#> 2 <NA> South Korea 3736 17 30#> 3 <NA> Italy 1694 34 83#> 4 Guangdong Mainland China 1349 7 1016#> 5 Henan Mainland China 1272 22 1198#> 6 Zhejiang Mainland China 1205 1 1046#> # … with 120 more rows, and 2 more variables:#> # latitude <dbl>, longitude <dbl>Why function
- Modularise the code to make it easier to understand the logic.
- Generalise the code to make it reusable later.
- Reduce the duplication and make it less error-prone.
copy and paste
covid19_csv1 <- read_covid19(urls[1])covid19_csv2 <- read_covid19(urls[2])covid19_csv3 <- read_covid19(urls[2])covid19_csv4 <- read_covid19(urls[4])covid19_csv5 <- read_covid19(urls[5])covid19_csv6 <- read_covid19(urls[6])covid19_csv6 <- read_covid19(urls[7])covid19_csv8 <- read_covid19(urls[8])covid19_csv9 <- read_covid19(urls[9])When it's gonna end? 😭
The goal of FP is to make it easy to express repeated actions using high-level verbs.
introduce typos/errors, when just simply copy and paste
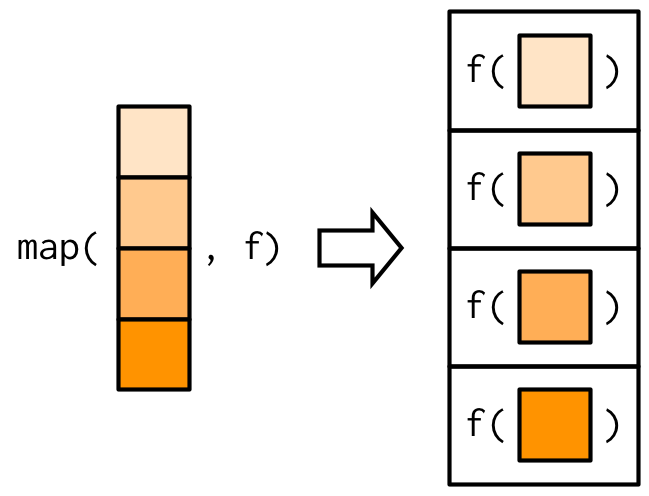
map(.x, .f, ...) for every element of .x, apply .f
- always returns a list
covid19_lst <- map(urls, read_covid19)covid19_lst[[length(covid19_lst)]]#> # A tibble: 3,980 x 13#> fips admin2 province_state country_region lat long#> <dbl> <chr> <chr> <chr> <dbl> <dbl>#> 1 NA <NA> <NA> Afghanistan 33.9 67.7 #> 2 NA <NA> <NA> Albania 41.2 20.2 #> 3 NA <NA> <NA> Algeria 28.0 1.66#> 4 NA <NA> <NA> Andorra 42.5 1.52#> 5 NA <NA> <NA> Angola -11.2 17.9 #> 6 NA <NA> <NA> Antigua and Barb… 17.1 -61.8 #> # … with 3,974 more rows, and 7 more variables:#> # confirmed <dbl>, deaths <dbl>, recovered <dbl>,#> # active <dbl>, combined_key <chr>, incident_rate <dbl>,#> # case_fatality_ratio <dbl>.f takes a function, formula, or vector.
- If a function, it is used as is.
- If a formula, e.g.
~ .x + 2, it is converted to a function. There are three ways to refer to the arguments:- For a single argument function, use
. - For a two argument function, use
.xand.y - For more arguments, use
..1,..2,..3etc
- For a single argument function, use
- If a character vector index by name and a numeric vector index by position.
covid19_lst <- map(urls, read_covid19)covid19_lst <- map(urls, ~ read_covid19(.x))covid19_lst <- map(urls, function(.x) read_covid19(.x))Bind multiple tibbles by row
bind_rows(covid19_lst)#> # A tibble: 1,069,141 x 16#> province_state country_region confirmed deaths recovered#> <chr> <chr> <dbl> <dbl> <dbl>#> 1 Hubei Mainland China 66907 2761 31536#> 2 <NA> South Korea 3736 17 30#> 3 <NA> Italy 1694 34 83#> 4 Guangdong Mainland China 1349 7 1016#> 5 Henan Mainland China 1272 22 1198#> 6 Zhejiang Mainland China 1205 1 1046#> # … with 1,069,135 more rows, and 11 more variables:#> # latitude <dbl>, longitude <dbl>, fips <dbl>,#> # admin2 <chr>, lat <dbl>, long <dbl>, active <dbl>,#> # combined_key <chr>, incidence_rate <dbl>,#> # case_fatality_ratio <dbl>, incident_rate <dbl>Hmm, where are the dates?
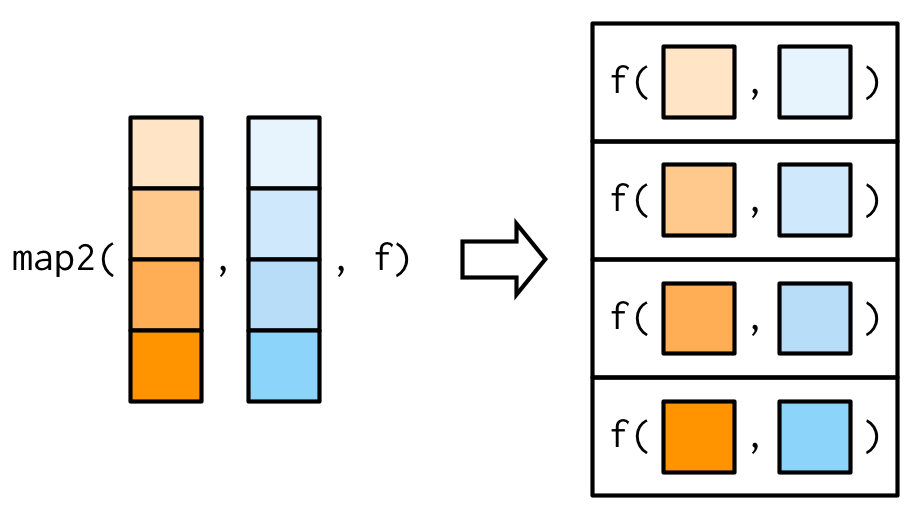
bind_rows(covid19_lst)#> # A tibble: 1,069,141 x 16#> province_state country_region confirmed deaths recovered#> <chr> <chr> <dbl> <dbl> <dbl>#> 1 Hubei Mainland China 66907 2761 31536#> 2 <NA> South Korea 3736 17 30#> 3 <NA> Italy 1694 34 83#> 4 Guangdong Mainland China 1349 7 1016#> 5 Henan Mainland China 1272 22 1198#> 6 Zhejiang Mainland China 1205 1 1046#> # … with 1,069,135 more rows, and 11 more variables:#> # latitude <dbl>, longitude <dbl>, fips <dbl>,#> # admin2 <chr>, lat <dbl>, long <dbl>, active <dbl>,#> # combined_key <chr>, incidence_rate <dbl>,#> # case_fatality_ratio <dbl>, incident_rate <dbl>map2(.x, .y, .f) for every element of .x and .y, apply .f
covid19_lst <- map2(urls, dates, ~ read_covid19(.x) %>% mutate(date = .y))bind_rows(covid19_lst)#> # A tibble: 1,069,141 x 17#> province_state country_region confirmed deaths recovered#> <chr> <chr> <dbl> <dbl> <dbl>#> 1 Hubei Mainland China 66907 2761 31536#> 2 <NA> South Korea 3736 17 30#> 3 <NA> Italy 1694 34 83#> 4 Guangdong Mainland China 1349 7 1016#> 5 Henan Mainland China 1272 22 1198#> 6 Zhejiang Mainland China 1205 1 1046#> # … with 1,069,135 more rows, and 12 more variables:#> # latitude <dbl>, longitude <dbl>, fips <dbl>,#> # admin2 <chr>, lat <dbl>, long <dbl>, active <dbl>,#> # combined_key <chr>, incidence_rate <dbl>,#> # case_fatality_ratio <dbl>, incident_rate <dbl>,#> # date <date>map_*() is a variant of map() that returns an output in the indicated type. map_dfr() returns data frames by row-binding.
covid19_raw <- map_dfr(urls, read_covid19, .id = "date")covid19_raw#> # A tibble: 1,069,141 x 17#> date province_state country_region confirmed deaths#> <chr> <chr> <chr> <dbl> <dbl>#> 1 03-01-2020 Hubei Mainland China 66907 2761#> 2 03-01-2020 <NA> South Korea 3736 17#> 3 03-01-2020 <NA> Italy 1694 34#> 4 03-01-2020 Guangdong Mainland China 1349 7#> 5 03-01-2020 Henan Mainland China 1272 22#> 6 03-01-2020 Zhejiang Mainland China 1205 1#> # … with 1,069,135 more rows, and 12 more variables:#> # recovered <dbl>, latitude <dbl>, longitude <dbl>,#> # fips <dbl>, admin2 <chr>, lat <dbl>, long <dbl>,#> # active <dbl>, combined_key <chr>, incidence_rate <dbl>,#> # case_fatality_ratio <dbl>, incident_rate <dbl>map() variants
map(.x, .f)maps over 1 argument returns a listmap_dbl(.x, .f)returns a double vectormap_chr(.x, .f)returns a character vectormap_lgl(.x, .f)returns a logical vectormap_dfr(.x, .f)returns a data frame created by row-bindingmap_dfc(.x, .f)returns a data frame created by col-bindingmap2_*(.x, .y, .f)maps over 2 argumentspmap_*(.l, .f)maps over > 2 arguments
Wrangle a bit
covid19_bystate <- covid19_raw %>% select(date, country_region:recovered) %>% mutate( date = mdy(date), country_region = case_when( country_region == "Korea, South" ~ "South Korea", country_region == "Mainland China" ~ "China", TRUE ~ country_region) )covid19_bystate#> # A tibble: 1,069,141 x 5#> date country_region confirmed deaths recovered#> <date> <chr> <dbl> <dbl> <dbl>#> 1 2020-03-01 China 66907 2761 31536#> 2 2020-03-01 South Korea 3736 17 30#> 3 2020-03-01 Italy 1694 34 83#> 4 2020-03-01 China 1349 7 1016#> 5 2020-03-01 China 1272 22 1198#> 6 2020-03-01 China 1205 1 1046#> # … with 1,069,135 more rowscovid19_byregion <- covid19_bystate %>% group_by(country_region, date) %>% summarise( confirmed = sum(confirmed, na.rm = TRUE), deaths = sum(deaths, na.rm = TRUE), recovered = sum(recovered, na.rm = TRUE) ) %>% mutate( new_confirmed = confirmed - lag(confirmed, na.rm = TRUE), new_deaths = deaths - lag(deaths, na.rm = TRUE), new_recovered = recovered - lag(recovered, na.rm = TRUE) ) %>% ungroup()Don't wanna type 😭, but I typed...
Apply a function across() multiple columns
covid19_byregion <- covid19_bystate %>% group_by(country_region, date) %>% summarise( across( confirmed:recovered, sum, na.rm = TRUE) ) %>% mutate( across( confirmed:recovered, ~ .x - lag(.x, na.rm = TRUE), .names = "new_{.col}") ) %>% ungroup()covid19_byregion <- covid19_bystate %>% group_by(country_region, date) %>% summarise( across( where(is.numeric), sum, na.rm = TRUE) ) %>% mutate( across( where(is.numeric), ~ .x - lag(.x, na.rm = TRUE), .names = "new_{.col}") ) %>% ungroup()covid19_byregion#> # A tibble: 56,107 x 8#> country_region date confirmed deaths recovered#> <chr> <date> <dbl> <dbl> <dbl>#> 1 Afghanistan 2020-03-01 1 0 0#> 2 Afghanistan 2020-03-02 1 0 0#> 3 Afghanistan 2020-03-03 2 0 0#> 4 Afghanistan 2020-03-04 4 0 0#> 5 Afghanistan 2020-03-05 4 0 0#> 6 Afghanistan 2020-03-06 4 0 0#> # … with 56,101 more rows, and 3 more variables:#> # new_confirmed <dbl>, new_deaths <dbl>,#> # new_recovered <dbl>nest() multiple columns into a list-column
covid19_lstcol <- covid19_byregion %>% nest(data = -country_region)covid19_lstcol#> # A tibble: 236 x 2#> country_region data #> <chr> <list> #> 1 Afghanistan <tibble[,7] [306 × 7]>#> 2 Albania <tibble[,7] [298 × 7]>#> 3 Algeria <tibble[,7] [306 × 7]>#> 4 Andorra <tibble[,7] [305 × 7]>#> 5 Angola <tibble[,7] [287 × 7]>#> 6 Antigua and Barbuda <tibble[,7] [294 × 7]>#> # … with 230 more rowsLook inside the list-column
covid19_au <- covid19_lstcol$data[[10]]covid19_au#> # A tibble: 306 x 7#> date confirmed deaths recovered new_confirmed#> <date> <dbl> <dbl> <dbl> <dbl>#> 1 2020-03-01 27 1 11 NA#> 2 2020-03-02 30 1 11 3#> 3 2020-03-03 39 1 11 9#> 4 2020-03-04 52 2 11 13#> 5 2020-03-05 55 2 21 3#> 6 2020-03-06 60 2 21 5#> # … with 300 more rows, and 2 more variables:#> # new_deaths <dbl>, new_recovered <dbl>Fit regression models and extract slopes
fit_au <- lm(new_recovered ~ lag(new_confirmed, n = 14), data = covid19_au)fit_au#> #> Call:#> lm(formula = new_recovered ~ lag(new_confirmed, n = 14), data = covid19_au)#> #> Coefficients:#> (Intercept) lag(new_confirmed, n = 14) #> 26.3548 0.5299slope_au <- coef(fit_au)[2]slope_au#> lag(new_confirmed, n = 14) #> 0.5299203If an error occured, safely() captures the error
- returns a named list of
resultanderror
safely_extract_slope <- safely(extract_slope, otherwise = NA_real_)covid19_lstcol$data %>% map(safely_extract_slope)#> [[1]]#> [[1]]$result#> lag(new_confirmed, n = 14) #> 0.6920523 #> #> [[1]]$error#> NULL#> #> #> [[2]]#> [[2]]$result#> lag(new_confirmed, n = 14) #> 0.6435756 #> #> [[2]]$error#> NULL#> #> #> [[3]]#> [[3]]$result#> lag(new_confirmed, n = 14) #> 0.5329582 #> #> [[3]]$error#> NULL#> #> #> [[4]]#> [[4]]$result#> lag(new_confirmed, n = 14) #> 0.5595175 #> #> [[4]]$error#> NULL#> #> #> [[5]]#> [[5]]$result#> lag(new_confirmed, n = 14) #> 0.5196389 #> #> [[5]]$error#> NULL#> #> #> [[6]]#> [[6]]$result#> lag(new_confirmed, n = 14) #> 0.7375993 #> #> [[6]]$error#> NULL#> #> #> [[7]]#> [[7]]$result#> lag(new_confirmed, n = 14) #> 0.8669078 #> #> [[7]]$error#> NULL#> #> #> [[8]]#> [[8]]$result#> lag(new_confirmed, n = 14) #> 0.8971555 #> #> [[8]]$error#> NULL#> #> #> [[9]]#> [[9]]$result#> [1] NA#> #> [[9]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[10]]#> [[10]]$result#> lag(new_confirmed, n = 14) #> 0.5299203 #> #> [[10]]$error#> NULL#> #> #> [[11]]#> [[11]]$result#> lag(new_confirmed, n = 14) #> 0.9182422 #> #> [[11]]$error#> NULL#> #> #> [[12]]#> [[12]]$result#> lag(new_confirmed, n = 14) #> 0.9642226 #> #> [[12]]$error#> NULL#> #> #> [[13]]#> [[13]]$result#> lag(new_confirmed, n = 14) #> 0.2903094 #> #> [[13]]$error#> NULL#> #> #> [[14]]#> [[14]]$result#> [1] NA#> #> [[14]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[15]]#> [[15]]$result#> lag(new_confirmed, n = 14) #> 0.8760548 #> #> [[15]]$error#> NULL#> #> #> [[16]]#> [[16]]$result#> lag(new_confirmed, n = 14) #> 0.8306039 #> #> [[16]]$error#> NULL#> #> #> [[17]]#> [[17]]$result#> lag(new_confirmed, n = 14) #> 0.1478383 #> #> [[17]]$error#> NULL#> #> #> [[18]]#> [[18]]$result#> lag(new_confirmed, n = 14) #> 1.038319 #> #> [[18]]$error#> NULL#> #> #> [[19]]#> [[19]]$result#> lag(new_confirmed, n = 14) #> -0.1417372 #> #> [[19]]$error#> NULL#> #> #> [[20]]#> [[20]]$result#> lag(new_confirmed, n = 14) #> 0.3760244 #> #> [[20]]$error#> NULL#> #> #> [[21]]#> [[21]]$result#> lag(new_confirmed, n = 14) #> 0.2665434 #> #> [[21]]$error#> NULL#> #> #> [[22]]#> [[22]]$result#> lag(new_confirmed, n = 14) #> 0.075549 #> #> [[22]]$error#> NULL#> #> #> [[23]]#> [[23]]$result#> lag(new_confirmed, n = 14) #> 0.6108023 #> #> [[23]]$error#> NULL#> #> #> [[24]]#> [[24]]$result#> lag(new_confirmed, n = 14) #> 0.6446241 #> #> [[24]]$error#> NULL#> #> #> [[25]]#> [[25]]$result#> lag(new_confirmed, n = 14) #> 0.491272 #> #> [[25]]$error#> NULL#> #> #> [[26]]#> [[26]]$result#> lag(new_confirmed, n = 14) #> 0.6540097 #> #> [[26]]$error#> NULL#> #> #> [[27]]#> [[27]]$result#> lag(new_confirmed, n = 14) #> 0.4138478 #> #> [[27]]$error#> NULL#> #> #> [[28]]#> [[28]]$result#> lag(new_confirmed, n = 14) #> 0.573295 #> #> [[28]]$error#> NULL#> #> #> [[29]]#> [[29]]$result#> lag(new_confirmed, n = 14) #> 0.6704973 #> #> [[29]]$error#> NULL#> #> #> [[30]]#> [[30]]$result#> lag(new_confirmed, n = 14) #> 0.8998348 #> #> [[30]]$error#> NULL#> #> #> [[31]]#> [[31]]$result#> lag(new_confirmed, n = 14) #> 0.05736442 #> #> [[31]]$error#> NULL#> #> #> [[32]]#> [[32]]$result#> lag(new_confirmed, n = 14) #> 0.5809416 #> #> [[32]]$error#> NULL#> #> #> [[33]]#> [[33]]$result#> lag(new_confirmed, n = 14) #> 0.1256359 #> #> [[33]]$error#> NULL#> #> #> [[34]]#> [[34]]$result#> lag(new_confirmed, n = 14) #> 0.1167159 #> #> [[34]]$error#> NULL#> #> #> [[35]]#> [[35]]$result#> lag(new_confirmed, n = 14) #> 0.9418862 #> #> [[35]]$error#> NULL#> #> #> [[36]]#> [[36]]$result#> [1] NA#> #> [[36]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[37]]#> [[37]]$result#> [1] NA#> #> [[37]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[38]]#> [[38]]$result#> lag(new_confirmed, n = 14) #> 0.2580678 #> #> [[38]]$error#> NULL#> #> #> [[39]]#> [[39]]$result#> lag(new_confirmed, n = 14) #> 0.4058524 #> #> [[39]]$error#> NULL#> #> #> [[40]]#> [[40]]$result#> [1] NA#> #> [[40]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[41]]#> [[41]]$result#> lag(new_confirmed, n = 14) #> 0.4110214 #> #> [[41]]$error#> NULL#> #> #> [[42]]#> [[42]]$result#> lag(new_confirmed, n = 14) #> 0.8410514 #> #> [[42]]$error#> NULL#> #> #> [[43]]#> [[43]]$result#> lag(new_confirmed, n = 14) #> 0.6855367 #> #> [[43]]$error#> NULL#> #> #> [[44]]#> [[44]]$result#> lag(new_confirmed, n = 14) #> 0.1374103 #> #> [[44]]$error#> NULL#> #> #> [[45]]#> [[45]]$result#> lag(new_confirmed, n = 14) #> 0.2689623 #> #> [[45]]$error#> NULL#> #> #> [[46]]#> [[46]]$result#> lag(new_confirmed, n = 14) #> 0.5826228 #> #> [[46]]$error#> NULL#> #> #> [[47]]#> [[47]]$result#> lag(new_confirmed, n = 14) #> 1.004124 #> #> [[47]]$error#> NULL#> #> #> [[48]]#> [[48]]$result#> lag(new_confirmed, n = 14) #> 0.6459742 #> #> [[48]]$error#> NULL#> #> #> [[49]]#> [[49]]$result#> lag(new_confirmed, n = 14) #> 0.8822818 #> #> [[49]]$error#> NULL#> #> #> [[50]]#> [[50]]$result#> [1] NA#> #> [[50]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[51]]#> [[51]]$result#> lag(new_confirmed, n = 14) #> 0.7600437 #> #> [[51]]$error#> NULL#> #> #> [[52]]#> [[52]]$result#> [1] NA#> #> [[52]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[53]]#> [[53]]$result#> lag(new_confirmed, n = 14) #> -0.01675946 #> #> [[53]]$error#> NULL#> #> #> [[54]]#> [[54]]$result#> [1] NA#> #> [[54]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[55]]#> [[55]]$result#> lag(new_confirmed, n = 14) #> 0.9635528 #> #> [[55]]$error#> NULL#> #> #> [[56]]#> [[56]]$result#> lag(new_confirmed, n = 14) #> 1.049798 #> #> [[56]]$error#> NULL#> #> #> [[57]]#> [[57]]$result#> lag(new_confirmed, n = 14) #> NA #> #> [[57]]$error#> NULL#> #> #> [[58]]#> [[58]]$result#> lag(new_confirmed, n = 14) #> 0.5009491 #> #> [[58]]$error#> NULL#> #> #> [[59]]#> [[59]]$result#> lag(new_confirmed, n = 14) #> 0.1780097 #> #> [[59]]$error#> NULL#> #> #> [[60]]#> [[60]]$result#> lag(new_confirmed, n = 14) #> 0.6290575 #> #> [[60]]$error#> NULL#> #> #> [[61]]#> [[61]]$result#> [1] NA#> #> [[61]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[62]]#> [[62]]$result#> lag(new_confirmed, n = 14) #> 0.05348175 #> #> [[62]]$error#> NULL#> #> #> [[63]]#> [[63]]$result#> lag(new_confirmed, n = 14) #> 0.2492007 #> #> [[63]]$error#> NULL#> #> #> [[64]]#> [[64]]$result#> lag(new_confirmed, n = 14) #> 0.5076202 #> #> [[64]]$error#> NULL#> #> #> [[65]]#> [[65]]$result#> lag(new_confirmed, n = 14) #> -0.01289268 #> #> [[65]]$error#> NULL#> #> #> [[66]]#> [[66]]$result#> lag(new_confirmed, n = 14) #> 0.1628999 #> #> [[66]]$error#> NULL#> #> #> [[67]]#> [[67]]$result#> lag(new_confirmed, n = 14) #> 0.8211096 #> #> [[67]]$error#> NULL#> #> #> [[68]]#> [[68]]$result#> lag(new_confirmed, n = 14) #> 0.6890107 #> #> [[68]]$error#> NULL#> #> #> [[69]]#> [[69]]$result#> lag(new_confirmed, n = 14) #> 0.4747831 #> #> [[69]]$error#> NULL#> #> #> [[70]]#> [[70]]$result#> [1] NA#> #> [[70]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[71]]#> [[71]]$result#> lag(new_confirmed, n = 14) #> 0.1370787 #> #> [[71]]$error#> NULL#> #> #> [[72]]#> [[72]]$result#> lag(new_confirmed, n = 14) #> 0.8498793 #> #> [[72]]$error#> NULL#> #> #> [[73]]#> [[73]]$result#> lag(new_confirmed, n = 14) #> 0.02613858 #> #> [[73]]$error#> NULL#> #> #> [[74]]#> [[74]]$result#> [1] NA#> #> [[74]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[75]]#> [[75]]$result#> lag(new_confirmed, n = 14) #> 0.4039257 #> #> [[75]]$error#> NULL#> #> #> [[76]]#> [[76]]$result#> lag(new_confirmed, n = 14) #> 0.2783185 #> #> [[76]]$error#> NULL#> #> #> [[77]]#> [[77]]$result#> [1] NA#> #> [[77]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[78]]#> [[78]]$result#> lag(new_confirmed, n = 14) #> 0.8992036 #> #> [[78]]$error#> NULL#> #> #> [[79]]#> [[79]]$result#> lag(new_confirmed, n = 14) #> 0.748486 #> #> [[79]]$error#> NULL#> #> #> [[80]]#> [[80]]$result#> lag(new_confirmed, n = 14) #> 0.4874482 #> #> [[80]]$error#> NULL#> #> #> [[81]]#> [[81]]$result#> [1] NA#> #> [[81]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[82]]#> [[82]]$result#> lag(new_confirmed, n = 14) #> 0.9351811 #> #> [[82]]$error#> NULL#> #> #> [[83]]#> [[83]]$result#> [1] NA#> #> [[83]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[84]]#> [[84]]$result#> lag(new_confirmed, n = 14) #> 0.02809673 #> #> [[84]]$error#> NULL#> #> #> [[85]]#> [[85]]$result#> [1] NA#> #> [[85]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[86]]#> [[86]]$result#> [1] NA#> #> [[86]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[87]]#> [[87]]$result#> lag(new_confirmed, n = 14) #> 0.8144507 #> #> [[87]]$error#> NULL#> #> #> [[88]]#> [[88]]$result#> [1] NA#> #> [[88]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[89]]#> [[89]]$result#> lag(new_confirmed, n = 14) #> 0.2512326 #> #> [[89]]$error#> NULL#> #> #> [[90]]#> [[90]]$result#> lag(new_confirmed, n = 14) #> 0.01763243 #> #> [[90]]$error#> NULL#> #> #> [[91]]#> [[91]]$result#> lag(new_confirmed, n = 14) #> 0.5299206 #> #> [[91]]$error#> NULL#> #> #> [[92]]#> [[92]]$result#> lag(new_confirmed, n = 14) #> 0.4224714 #> #> [[92]]$error#> NULL#> #> #> [[93]]#> [[93]]$result#> lag(new_confirmed, n = 14) #> -0.01196319 #> #> [[93]]$error#> NULL#> #> #> [[94]]#> [[94]]$result#> lag(new_confirmed, n = 14) #> 0.2954188 #> #> [[94]]$error#> NULL#> #> #> [[95]]#> [[95]]$result#> [1] NA#> #> [[95]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[96]]#> [[96]]$result#> [1] NA#> #> [[96]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[97]]#> [[97]]$result#> lag(new_confirmed, n = 14) #> 0.4821869 #> #> [[97]]$error#> NULL#> #> #> [[98]]#> [[98]]$result#> lag(new_confirmed, n = 14) #> 0.7901768 #> #> [[98]]$error#> NULL#> #> #> [[99]]#> [[99]]$result#> lag(new_confirmed, n = 14) #> 0.8979504 #> #> [[99]]$error#> NULL#> #> #> [[100]]#> [[100]]$result#> lag(new_confirmed, n = 14) #> 0.9487975 #> #> [[100]]$error#> NULL#> #> #> [[101]]#> [[101]]$result#> lag(new_confirmed, n = 14) #> 0.7877892 #> #> [[101]]$error#> NULL#> #> #> [[102]]#> [[102]]$result#> [1] NA#> #> [[102]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[103]]#> [[103]]$result#> lag(new_confirmed, n = 14) #> 0.8682548 #> #> [[103]]$error#> NULL#> #> #> [[104]]#> [[104]]$result#> lag(new_confirmed, n = 14) #> 0.2087615 #> #> [[104]]$error#> NULL#> #> #> [[105]]#> [[105]]$result#> lag(new_confirmed, n = 14) #> 0.8144906 #> #> [[105]]$error#> NULL#> #> #> [[106]]#> [[106]]$result#> lag(new_confirmed, n = 14) #> 0.7282967 #> #> [[106]]$error#> NULL#> #> #> [[107]]#> [[107]]$result#> lag(new_confirmed, n = 14) #> 0.5431636 #> #> [[107]]$error#> NULL#> #> #> [[108]]#> [[108]]$result#> lag(new_confirmed, n = 14) #> 0.8678327 #> #> [[108]]$error#> NULL#> #> #> [[109]]#> [[109]]$result#> [1] NA#> #> [[109]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[110]]#> [[110]]$result#> lag(new_confirmed, n = 14) #> 1.177228 #> #> [[110]]$error#> NULL#> #> #> [[111]]#> [[111]]$result#> lag(new_confirmed, n = 14) #> 0.1624997 #> #> [[111]]$error#> NULL#> #> #> [[112]]#> [[112]]$result#> lag(new_confirmed, n = 14) #> 0.7411408 #> #> [[112]]$error#> NULL#> #> #> [[113]]#> [[113]]$result#> lag(new_confirmed, n = 14) #> 0.02179744 #> #> [[113]]$error#> NULL#> #> #> [[114]]#> [[114]]$result#> lag(new_confirmed, n = 14) #> 0.9123329 #> #> [[114]]$error#> NULL#> #> #> [[115]]#> [[115]]$result#> lag(new_confirmed, n = 14) #> 0.1588501 #> #> [[115]]$error#> NULL#> #> #> [[116]]#> [[116]]$result#> lag(new_confirmed, n = 14) #> 0.0223889 #> #> [[116]]$error#> NULL#> #> #> [[117]]#> [[117]]$result#> lag(new_confirmed, n = 14) #> 0.9392607 #> #> [[117]]$error#> NULL#> #> #> [[118]]#> [[118]]$result#> lag(new_confirmed, n = 14) #> 0.7137065 #> #> [[118]]$error#> NULL#> #> #> [[119]]#> [[119]]$result#> lag(new_confirmed, n = 14) #> 0.1059572 #> #> [[119]]$error#> NULL#> #> #> [[120]]#> [[120]]$result#> lag(new_confirmed, n = 14) #> 0.1280151 #> #> [[120]]$error#> NULL#> #> #> [[121]]#> [[121]]$result#> lag(new_confirmed, n = 14) #> 0.7095778 #> #> [[121]]$error#> NULL#> #> #> [[122]]#> [[122]]$result#> lag(new_confirmed, n = 14) #> 0.8902059 #> #> [[122]]$error#> NULL#> #> #> [[123]]#> [[123]]$result#> lag(new_confirmed, n = 14) #> 0.8601565 #> #> [[123]]$error#> NULL#> #> #> [[124]]#> [[124]]$result#> lag(new_confirmed, n = 14) #> 0.933333 #> #> [[124]]$error#> NULL#> #> #> [[125]]#> [[125]]$result#> [1] NA#> #> [[125]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[126]]#> [[126]]$result#> [1] NA#> #> [[126]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[127]]#> [[127]]$result#> lag(new_confirmed, n = 14) #> 0.7684239 #> #> [[127]]$error#> NULL#> #> #> [[128]]#> [[128]]$result#> lag(new_confirmed, n = 14) #> 0.2407153 #> #> [[128]]$error#> NULL#> #> #> [[129]]#> [[129]]$result#> lag(new_confirmed, n = 14) #> 0.8741184 #> #> [[129]]$error#> NULL#> #> #> [[130]]#> [[130]]$result#> lag(new_confirmed, n = 14) #> 0.8370697 #> #> [[130]]$error#> NULL#> #> #> [[131]]#> [[131]]$result#> lag(new_confirmed, n = 14) #> 0.4458898 #> #> [[131]]$error#> NULL#> #> #> [[132]]#> [[132]]$result#> lag(new_confirmed, n = 14) #> 0.8722886 #> #> [[132]]$error#> NULL#> #> #> [[133]]#> [[133]]$result#> lag(new_confirmed, n = 14) #> -0.01612903 #> #> [[133]]$error#> NULL#> #> #> [[134]]#> [[134]]$result#> [1] NA#> #> [[134]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[135]]#> [[135]]$result#> lag(new_confirmed, n = 14) #> 0.7781136 #> #> [[135]]$error#> NULL#> #> #> [[136]]#> [[136]]$result#> lag(new_confirmed, n = 14) #> 0.4789353 #> #> [[136]]$error#> NULL#> #> #> [[137]]#> [[137]]$result#> [1] NA#> #> [[137]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[138]]#> [[138]]$result#> lag(new_confirmed, n = 14) #> 0.5295625 #> #> [[138]]$error#> NULL#> #> #> [[139]]#> [[139]]$result#> lag(new_confirmed, n = 14) #> 0.8784718 #> #> [[139]]$error#> NULL#> #> #> [[140]]#> [[140]]$result#> lag(new_confirmed, n = 14) #> 0.4523111 #> #> [[140]]$error#> NULL#> #> #> [[141]]#> [[141]]$result#> lag(new_confirmed, n = 14) #> 0.06500141 #> #> [[141]]$error#> NULL#> #> #> [[142]]#> [[142]]$result#> lag(new_confirmed, n = 14) #> 0.7416119 #> #> [[142]]$error#> NULL#> #> #> [[143]]#> [[143]]$result#> lag(new_confirmed, n = 14) #> 0.8947562 #> #> [[143]]$error#> NULL#> #> #> [[144]]#> [[144]]$result#> lag(new_confirmed, n = 14) #> 0.7780699 #> #> [[144]]$error#> NULL#> #> #> [[145]]#> [[145]]$result#> lag(new_confirmed, n = 14) #> -0.003802281 #> #> [[145]]$error#> NULL#> #> #> [[146]]#> [[146]]$result#> lag(new_confirmed, n = 14) #> 1.044826 #> #> [[146]]$error#> NULL#> #> #> [[147]]#> [[147]]$result#> lag(new_confirmed, n = 14) #> 0.9334443 #> #> [[147]]$error#> NULL#> #> #> [[148]]#> [[148]]$result#> lag(new_confirmed, n = 14) #> 0.005462774 #> #> [[148]]$error#> NULL#> #> #> [[149]]#> [[149]]$result#> lag(new_confirmed, n = 14) #> 0.7127593 #> #> [[149]]$error#> NULL#> #> #> [[150]]#> [[150]]$result#> lag(new_confirmed, n = 14) #> 0.6934141 #> #> [[150]]$error#> NULL#> #> #> [[151]]#> [[151]]$result#> lag(new_confirmed, n = 14) #> 0.4480632 #> #> [[151]]$error#> NULL#> #> #> [[152]]#> [[152]]$result#> lag(new_confirmed, n = 14) #> 0.7526479 #> #> [[152]]$error#> NULL#> #> #> [[153]]#> [[153]]$result#> lag(new_confirmed, n = 14) #> 0.6662816 #> #> [[153]]$error#> NULL#> #> #> [[154]]#> [[154]]$result#> lag(new_confirmed, n = 14) #> 0.2028236 #> #> [[154]]$error#> NULL#> #> #> [[155]]#> [[155]]$result#> [1] NA#> #> [[155]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[156]]#> [[156]]$result#> lag(new_confirmed, n = 14) #> 0.9219172 #> #> [[156]]$error#> NULL#> #> #> [[157]]#> [[157]]$result#> [1] NA#> #> [[157]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[158]]#> [[158]]$result#> lag(new_confirmed, n = 14) #> 0.5813632 #> #> [[158]]$error#> NULL#> #> #> [[159]]#> [[159]]$result#> [1] NA#> #> [[159]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[160]]#> [[160]]$result#> lag(new_confirmed, n = 14) #> 0.7849124 #> #> [[160]]$error#> NULL#> #> #> [[161]]#> [[161]]$result#> lag(new_confirmed, n = 14) #> 0.0817926 #> #> [[161]]$error#> NULL#> #> #> [[162]]#> [[162]]$result#> lag(new_confirmed, n = 14) #> 0.7567328 #> #> [[162]]$error#> NULL#> #> #> [[163]]#> [[163]]$result#> lag(new_confirmed, n = 14) #> 0.4486768 #> #> [[163]]$error#> NULL#> #> #> [[164]]#> [[164]]$result#> lag(new_confirmed, n = 14) #> 0.9711064 #> #> [[164]]$error#> NULL#> #> #> [[165]]#> [[165]]$result#> lag(new_confirmed, n = 14) #> 0.8404479 #> #> [[165]]$error#> NULL#> #> #> [[166]]#> [[166]]$result#> lag(new_confirmed, n = 14) #> 0.8340365 #> #> [[166]]$error#> NULL#> #> #> [[167]]#> [[167]]$result#> [1] NA#> #> [[167]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[168]]#> [[168]]$result#> lag(new_confirmed, n = 14) #> 1.146428 #> #> [[168]]$error#> NULL#> #> #> [[169]]#> [[169]]$result#> [1] NA#> #> [[169]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[170]]#> [[170]]$result#> [1] NA#> #> [[170]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[171]]#> [[171]]$result#> [1] NA#> #> [[171]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[172]]#> [[172]]$result#> [1] NA#> #> [[172]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[173]]#> [[173]]$result#> [1] NA#> #> [[173]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[174]]#> [[174]]$result#> lag(new_confirmed, n = 14) #> 0.9711078 #> #> [[174]]$error#> NULL#> #> #> [[175]]#> [[175]]$result#> lag(new_confirmed, n = 14) #> 0.9645341 #> #> [[175]]$error#> NULL#> #> #> [[176]]#> [[176]]$result#> [1] NA#> #> [[176]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[177]]#> [[177]]$result#> lag(new_confirmed, n = 14) #> 0.1992464 #> #> [[177]]$error#> NULL#> #> #> [[178]]#> [[178]]$result#> [1] NA#> #> [[178]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[179]]#> [[179]]$result#> lag(new_confirmed, n = 14) #> 0.05725861 #> #> [[179]]$error#> NULL#> #> #> [[180]]#> [[180]]$result#> lag(new_confirmed, n = 14) #> 0.5481366 #> #> [[180]]$error#> NULL#> #> #> [[181]]#> [[181]]$result#> [1] NA#> #> [[181]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[182]]#> [[182]]$result#> lag(new_confirmed, n = 14) #> -0.05410779 #> #> [[182]]$error#> NULL#> #> #> [[183]]#> [[183]]$result#> lag(new_confirmed, n = 14) #> -0.007462687 #> #> [[183]]$error#> NULL#> #> #> [[184]]#> [[184]]$result#> lag(new_confirmed, n = 14) #> 0.3680185 #> #> [[184]]$error#> NULL#> #> #> [[185]]#> [[185]]$result#> lag(new_confirmed, n = 14) #> -0.00138019 #> #> [[185]]$error#> NULL#> #> #> [[186]]#> [[186]]$result#> lag(new_confirmed, n = 14) #> 0.8943568 #> #> [[186]]$error#> NULL#> #> #> [[187]]#> [[187]]$result#> lag(new_confirmed, n = 14) #> 0.4699798 #> #> [[187]]$error#> NULL#> #> #> [[188]]#> [[188]]$result#> lag(new_confirmed, n = 14) #> -0.001714412 #> #> [[188]]$error#> NULL#> #> #> [[189]]#> [[189]]$result#> lag(new_confirmed, n = 14) #> -0.01177239 #> #> [[189]]$error#> NULL#> #> #> [[190]]#> [[190]]$result#> lag(new_confirmed, n = 14) #> 0.4459296 #> #> [[190]]$error#> NULL#> #> #> [[191]]#> [[191]]$result#> lag(new_confirmed, n = 14) #> 0.741075 #> #> [[191]]$error#> NULL#> #> #> [[192]]#> [[192]]$result#> lag(new_confirmed, n = 14) #> 0.7394529 #> #> [[192]]$error#> NULL#> #> #> [[193]]#> [[193]]$result#> lag(new_confirmed, n = 14) #> 0.9754129 #> #> [[193]]$error#> NULL#> #> #> [[194]]#> [[194]]$result#> lag(new_confirmed, n = 14) #> -0.004582951 #> #> [[194]]$error#> NULL#> #> #> [[195]]#> [[195]]$result#> lag(new_confirmed, n = 14) #> 0.1134682 #> #> [[195]]$error#> NULL#> #> #> [[196]]#> [[196]]$result#> lag(new_confirmed, n = 14) #> 0.8579614 #> #> [[196]]$error#> NULL#> #> #> [[197]]#> [[197]]$result#> lag(new_confirmed, n = 14) #> 0.7022731 #> #> [[197]]$error#> NULL#> #> #> [[198]]#> [[198]]$result#> lag(new_confirmed, n = 14) #> 0.0003349323 #> #> [[198]]$error#> NULL#> #> #> [[199]]#> [[199]]$result#> lag(new_confirmed, n = 14) #> -0.01591102 #> #> [[199]]$error#> NULL#> #> #> [[200]]#> [[200]]$result#> lag(new_confirmed, n = 14) #> 0.902218 #> #> [[200]]$error#> NULL#> #> #> [[201]]#> [[201]]$result#> [1] NA#> #> [[201]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[202]]#> [[202]]$result#> lag(new_confirmed, n = 14) #> 0.2367338 #> #> [[202]]$error#> NULL#> #> #> [[203]]#> [[203]]$result#> lag(new_confirmed, n = 14) #> 0.6937724 #> #> [[203]]$error#> NULL#> #> #> [[204]]#> [[204]]$result#> lag(new_confirmed, n = 14) #> 8.226172e-05 #> #> [[204]]$error#> NULL#> #> #> [[205]]#> [[205]]$result#> lag(new_confirmed, n = 14) #> 0.7049068 #> #> [[205]]$error#> NULL#> #> #> [[206]]#> [[206]]$result#> lag(new_confirmed, n = 14) #> 0.4773192 #> #> [[206]]$error#> NULL#> #> #> [[207]]#> [[207]]$result#> [1] NA#> #> [[207]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[208]]#> [[208]]$result#> [1] NA#> #> [[208]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[209]]#> [[209]]$result#> lag(new_confirmed, n = 14) #> 0.2683079 #> #> [[209]]$error#> NULL#> #> #> [[210]]#> [[210]]$result#> lag(new_confirmed, n = 14) #> 0.3773631 #> #> [[210]]$error#> NULL#> #> #> [[211]]#> [[211]]$result#> lag(new_confirmed, n = 14) #> 0.09471127 #> #> [[211]]$error#> NULL#> #> #> [[212]]#> [[212]]$result#> lag(new_confirmed, n = 14) #> 0.791494 #> #> [[212]]$error#> NULL#> #> #> [[213]]#> [[213]]$result#> [1] NA#> #> [[213]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[214]]#> [[214]]$result#> [1] NA#> #> [[214]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[215]]#> [[215]]$result#> lag(new_confirmed, n = 14) #> 0.9127807 #> #> [[215]]$error#> NULL#> #> #> [[216]]#> [[216]]$result#> lag(new_confirmed, n = 14) #> 0.6683978 #> #> [[216]]$error#> NULL#> #> #> [[217]]#> [[217]]$result#> lag(new_confirmed, n = 14) #> 0.4777528 #> #> [[217]]$error#> NULL#> #> #> [[218]]#> [[218]]$result#> lag(new_confirmed, n = 14) #> 0.289256 #> #> [[218]]$error#> NULL#> #> #> [[219]]#> [[219]]$result#> lag(new_confirmed, n = 14) #> 0.09288966 #> #> [[219]]$error#> NULL#> #> #> [[220]]#> [[220]]$result#> lag(new_confirmed, n = 14) #> 0.1472352 #> #> [[220]]$error#> NULL#> #> #> [[221]]#> [[221]]$result#> [1] NA#> #> [[221]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[222]]#> [[222]]$result#> lag(new_confirmed, n = 14) #> 0.8942314 #> #> [[222]]$error#> NULL#> #> #> [[223]]#> [[223]]$result#> lag(new_confirmed, n = 14) #> 0.8124493 #> #> [[223]]$error#> NULL#> #> #> [[224]]#> [[224]]$result#> lag(new_confirmed, n = 14) #> 0.001282686 #> #> [[224]]$error#> NULL#> #> #> [[225]]#> [[225]]$result#> lag(new_confirmed, n = 14) #> 1.164049 #> #> [[225]]$error#> NULL#> #> #> [[226]]#> [[226]]$result#> lag(new_confirmed, n = 14) #> -0.7243288 #> #> [[226]]$error#> NULL#> #> #> [[227]]#> [[227]]$result#> lag(new_confirmed, n = 14) #> 0.9300546 #> #> [[227]]$error#> NULL#> #> #> [[228]]#> [[228]]$result#> lag(new_confirmed, n = 14) #> NA #> #> [[228]]$error#> NULL#> #> #> [[229]]#> [[229]]$result#> [1] NA#> #> [[229]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[230]]#> [[230]]$result#> lag(new_confirmed, n = 14) #> 0.9314963 #> #> [[230]]$error#> NULL#> #> #> [[231]]#> [[231]]$result#> [1] NA#> #> [[231]]$error#> <simpleError in lm.fit(x, y, offset = offset, singular.ok = singular.ok, ...): 0 (non-NA) cases>#> #> #> [[232]]#> [[232]]$result#> lag(new_confirmed, n = 14) #> 0.3004533 #> #> [[232]]$error#> NULL#> #> #> [[233]]#> [[233]]$result#> lag(new_confirmed, n = 14) #> 0.9587314 #> #> [[233]]$error#> NULL#> #> #> [[234]]#> [[234]]$result#> lag(new_confirmed, n = 14) #> 0.2780126 #> #> [[234]]$error#> NULL#> #> #> [[235]]#> [[235]]$result#> lag(new_confirmed, n = 14) #> 0.4839777 #> #> [[235]]$error#> NULL#> #> #> [[236]]#> [[236]]$result#> lag(new_confirmed, n = 14) #> 0.4202595 #> #> [[236]]$error#> NULLPull out results
covid19_lstcol$data %>% map(safely_extract_slope) %>% map("result")#> [[1]]#> lag(new_confirmed, n = 14) #> 0.6920523 #> #> [[2]]#> lag(new_confirmed, n = 14) #> 0.6435756 #> #> [[3]]#> lag(new_confirmed, n = 14) #> 0.5329582 #> #> [[4]]#> lag(new_confirmed, n = 14) #> 0.5595175 #> #> [[5]]#> lag(new_confirmed, n = 14) #> 0.5196389 #> #> [[6]]#> lag(new_confirmed, n = 14) #> 0.7375993 #> #> [[7]]#> lag(new_confirmed, n = 14) #> 0.8669078 #> #> [[8]]#> lag(new_confirmed, n = 14) #> 0.8971555 #> #> [[9]]#> [1] NA#> #> [[10]]#> lag(new_confirmed, n = 14) #> 0.5299203 #> #> [[11]]#> lag(new_confirmed, n = 14) #> 0.9182422 #> #> [[12]]#> lag(new_confirmed, n = 14) #> 0.9642226 #> #> [[13]]#> lag(new_confirmed, n = 14) #> 0.2903094 #> #> [[14]]#> [1] NA#> #> [[15]]#> lag(new_confirmed, n = 14) #> 0.8760548 #> #> [[16]]#> lag(new_confirmed, n = 14) #> 0.8306039 #> #> [[17]]#> lag(new_confirmed, n = 14) #> 0.1478383 #> #> [[18]]#> lag(new_confirmed, n = 14) #> 1.038319 #> #> [[19]]#> lag(new_confirmed, n = 14) #> -0.1417372 #> #> [[20]]#> lag(new_confirmed, n = 14) #> 0.3760244 #> #> [[21]]#> lag(new_confirmed, n = 14) #> 0.2665434 #> #> [[22]]#> lag(new_confirmed, n = 14) #> 0.075549 #> #> [[23]]#> lag(new_confirmed, n = 14) #> 0.6108023 #> #> [[24]]#> lag(new_confirmed, n = 14) #> 0.6446241 #> #> [[25]]#> lag(new_confirmed, n = 14) #> 0.491272 #> #> [[26]]#> lag(new_confirmed, n = 14) #> 0.6540097 #> #> [[27]]#> lag(new_confirmed, n = 14) #> 0.4138478 #> #> [[28]]#> lag(new_confirmed, n = 14) #> 0.573295 #> #> [[29]]#> lag(new_confirmed, n = 14) #> 0.6704973 #> #> [[30]]#> lag(new_confirmed, n = 14) #> 0.8998348 #> #> [[31]]#> lag(new_confirmed, n = 14) #> 0.05736442 #> #> [[32]]#> lag(new_confirmed, n = 14) #> 0.5809416 #> #> [[33]]#> lag(new_confirmed, n = 14) #> 0.1256359 #> #> [[34]]#> lag(new_confirmed, n = 14) #> 0.1167159 #> #> [[35]]#> lag(new_confirmed, n = 14) #> 0.9418862 #> #> [[36]]#> [1] NA#> #> [[37]]#> [1] NA#> #> [[38]]#> lag(new_confirmed, n = 14) #> 0.2580678 #> #> [[39]]#> lag(new_confirmed, n = 14) #> 0.4058524 #> #> [[40]]#> [1] NA#> #> [[41]]#> lag(new_confirmed, n = 14) #> 0.4110214 #> #> [[42]]#> lag(new_confirmed, n = 14) #> 0.8410514 #> #> [[43]]#> lag(new_confirmed, n = 14) #> 0.6855367 #> #> [[44]]#> lag(new_confirmed, n = 14) #> 0.1374103 #> #> [[45]]#> lag(new_confirmed, n = 14) #> 0.2689623 #> #> [[46]]#> lag(new_confirmed, n = 14) #> 0.5826228 #> #> [[47]]#> lag(new_confirmed, n = 14) #> 1.004124 #> #> [[48]]#> lag(new_confirmed, n = 14) #> 0.6459742 #> #> [[49]]#> lag(new_confirmed, n = 14) #> 0.8822818 #> #> [[50]]#> [1] NA#> #> [[51]]#> lag(new_confirmed, n = 14) #> 0.7600437 #> #> [[52]]#> [1] NA#> #> [[53]]#> lag(new_confirmed, n = 14) #> -0.01675946 #> #> [[54]]#> [1] NA#> #> [[55]]#> lag(new_confirmed, n = 14) #> 0.9635528 #> #> [[56]]#> lag(new_confirmed, n = 14) #> 1.049798 #> #> [[57]]#> lag(new_confirmed, n = 14) #> NA #> #> [[58]]#> lag(new_confirmed, n = 14) #> 0.5009491 #> #> [[59]]#> lag(new_confirmed, n = 14) #> 0.1780097 #> #> [[60]]#> lag(new_confirmed, n = 14) #> 0.6290575 #> #> [[61]]#> [1] NA#> #> [[62]]#> lag(new_confirmed, n = 14) #> 0.05348175 #> #> [[63]]#> lag(new_confirmed, n = 14) #> 0.2492007 #> #> [[64]]#> lag(new_confirmed, n = 14) #> 0.5076202 #> #> [[65]]#> lag(new_confirmed, n = 14) #> -0.01289268 #> #> [[66]]#> lag(new_confirmed, n = 14) #> 0.1628999 #> #> [[67]]#> lag(new_confirmed, n = 14) #> 0.8211096 #> #> [[68]]#> lag(new_confirmed, n = 14) #> 0.6890107 #> #> [[69]]#> lag(new_confirmed, n = 14) #> 0.4747831 #> #> [[70]]#> [1] NA#> #> [[71]]#> lag(new_confirmed, n = 14) #> 0.1370787 #> #> [[72]]#> lag(new_confirmed, n = 14) #> 0.8498793 #> #> [[73]]#> lag(new_confirmed, n = 14) #> 0.02613858 #> #> [[74]]#> [1] NA#> #> [[75]]#> lag(new_confirmed, n = 14) #> 0.4039257 #> #> [[76]]#> lag(new_confirmed, n = 14) #> 0.2783185 #> #> [[77]]#> [1] NA#> #> [[78]]#> lag(new_confirmed, n = 14) #> 0.8992036 #> #> [[79]]#> lag(new_confirmed, n = 14) #> 0.748486 #> #> [[80]]#> lag(new_confirmed, n = 14) #> 0.4874482 #> #> [[81]]#> [1] NA#> #> [[82]]#> lag(new_confirmed, n = 14) #> 0.9351811 #> #> [[83]]#> [1] NA#> #> [[84]]#> lag(new_confirmed, n = 14) #> 0.02809673 #> #> [[85]]#> [1] NA#> #> [[86]]#> [1] NA#> #> [[87]]#> lag(new_confirmed, n = 14) #> 0.8144507 #> #> [[88]]#> [1] NA#> #> [[89]]#> lag(new_confirmed, n = 14) #> 0.2512326 #> #> [[90]]#> lag(new_confirmed, n = 14) #> 0.01763243 #> #> [[91]]#> lag(new_confirmed, n = 14) #> 0.5299206 #> #> [[92]]#> lag(new_confirmed, n = 14) #> 0.4224714 #> #> [[93]]#> lag(new_confirmed, n = 14) #> -0.01196319 #> #> [[94]]#> lag(new_confirmed, n = 14) #> 0.2954188 #> #> [[95]]#> [1] NA#> #> [[96]]#> [1] NA#> #> [[97]]#> lag(new_confirmed, n = 14) #> 0.4821869 #> #> [[98]]#> lag(new_confirmed, n = 14) #> 0.7901768 #> #> [[99]]#> lag(new_confirmed, n = 14) #> 0.8979504 #> #> [[100]]#> lag(new_confirmed, n = 14) #> 0.9487975 #> #> [[101]]#> lag(new_confirmed, n = 14) #> 0.7877892 #> #> [[102]]#> [1] NA#> #> [[103]]#> lag(new_confirmed, n = 14) #> 0.8682548 #> #> [[104]]#> lag(new_confirmed, n = 14) #> 0.2087615 #> #> [[105]]#> lag(new_confirmed, n = 14) #> 0.8144906 #> #> [[106]]#> lag(new_confirmed, n = 14) #> 0.7282967 #> #> [[107]]#> lag(new_confirmed, n = 14) #> 0.5431636 #> #> [[108]]#> lag(new_confirmed, n = 14) #> 0.8678327 #> #> [[109]]#> [1] NA#> #> [[110]]#> lag(new_confirmed, n = 14) #> 1.177228 #> #> [[111]]#> lag(new_confirmed, n = 14) #> 0.1624997 #> #> [[112]]#> lag(new_confirmed, n = 14) #> 0.7411408 #> #> [[113]]#> lag(new_confirmed, n = 14) #> 0.02179744 #> #> [[114]]#> lag(new_confirmed, n = 14) #> 0.9123329 #> #> [[115]]#> lag(new_confirmed, n = 14) #> 0.1588501 #> #> [[116]]#> lag(new_confirmed, n = 14) #> 0.0223889 #> #> [[117]]#> lag(new_confirmed, n = 14) #> 0.9392607 #> #> [[118]]#> lag(new_confirmed, n = 14) #> 0.7137065 #> #> [[119]]#> lag(new_confirmed, n = 14) #> 0.1059572 #> #> [[120]]#> lag(new_confirmed, n = 14) #> 0.1280151 #> #> [[121]]#> lag(new_confirmed, n = 14) #> 0.7095778 #> #> [[122]]#> lag(new_confirmed, n = 14) #> 0.8902059 #> #> [[123]]#> lag(new_confirmed, n = 14) #> 0.8601565 #> #> [[124]]#> lag(new_confirmed, n = 14) #> 0.933333 #> #> [[125]]#> [1] NA#> #> [[126]]#> [1] NA#> #> [[127]]#> lag(new_confirmed, n = 14) #> 0.7684239 #> #> [[128]]#> lag(new_confirmed, n = 14) #> 0.2407153 #> #> [[129]]#> lag(new_confirmed, n = 14) #> 0.8741184 #> #> [[130]]#> lag(new_confirmed, n = 14) #> 0.8370697 #> #> [[131]]#> lag(new_confirmed, n = 14) #> 0.4458898 #> #> [[132]]#> lag(new_confirmed, n = 14) #> 0.8722886 #> #> [[133]]#> lag(new_confirmed, n = 14) #> -0.01612903 #> #> [[134]]#> [1] NA#> #> [[135]]#> lag(new_confirmed, n = 14) #> 0.7781136 #> #> [[136]]#> lag(new_confirmed, n = 14) #> 0.4789353 #> #> [[137]]#> [1] NA#> #> [[138]]#> lag(new_confirmed, n = 14) #> 0.5295625 #> #> [[139]]#> lag(new_confirmed, n = 14) #> 0.8784718 #> #> [[140]]#> lag(new_confirmed, n = 14) #> 0.4523111 #> #> [[141]]#> lag(new_confirmed, n = 14) #> 0.06500141 #> #> [[142]]#> lag(new_confirmed, n = 14) #> 0.7416119 #> #> [[143]]#> lag(new_confirmed, n = 14) #> 0.8947562 #> #> [[144]]#> lag(new_confirmed, n = 14) #> 0.7780699 #> #> [[145]]#> lag(new_confirmed, n = 14) #> -0.003802281 #> #> [[146]]#> lag(new_confirmed, n = 14) #> 1.044826 #> #> [[147]]#> lag(new_confirmed, n = 14) #> 0.9334443 #> #> [[148]]#> lag(new_confirmed, n = 14) #> 0.005462774 #> #> [[149]]#> lag(new_confirmed, n = 14) #> 0.7127593 #> #> [[150]]#> lag(new_confirmed, n = 14) #> 0.6934141 #> #> [[151]]#> lag(new_confirmed, n = 14) #> 0.4480632 #> #> [[152]]#> lag(new_confirmed, n = 14) #> 0.7526479 #> #> [[153]]#> lag(new_confirmed, n = 14) #> 0.6662816 #> #> [[154]]#> lag(new_confirmed, n = 14) #> 0.2028236 #> #> [[155]]#> [1] NA#> #> [[156]]#> lag(new_confirmed, n = 14) #> 0.9219172 #> #> [[157]]#> [1] NA#> #> [[158]]#> lag(new_confirmed, n = 14) #> 0.5813632 #> #> [[159]]#> [1] NA#> #> [[160]]#> lag(new_confirmed, n = 14) #> 0.7849124 #> #> [[161]]#> lag(new_confirmed, n = 14) #> 0.0817926 #> #> [[162]]#> lag(new_confirmed, n = 14) #> 0.7567328 #> #> [[163]]#> lag(new_confirmed, n = 14) #> 0.4486768 #> #> [[164]]#> lag(new_confirmed, n = 14) #> 0.9711064 #> #> [[165]]#> lag(new_confirmed, n = 14) #> 0.8404479 #> #> [[166]]#> lag(new_confirmed, n = 14) #> 0.8340365 #> #> [[167]]#> [1] NA#> #> [[168]]#> lag(new_confirmed, n = 14) #> 1.146428 #> #> [[169]]#> [1] NA#> #> [[170]]#> [1] NA#> #> [[171]]#> [1] NA#> #> [[172]]#> [1] NA#> #> [[173]]#> [1] NA#> #> [[174]]#> lag(new_confirmed, n = 14) #> 0.9711078 #> #> [[175]]#> lag(new_confirmed, n = 14) #> 0.9645341 #> #> [[176]]#> [1] NA#> #> [[177]]#> lag(new_confirmed, n = 14) #> 0.1992464 #> #> [[178]]#> [1] NA#> #> [[179]]#> lag(new_confirmed, n = 14) #> 0.05725861 #> #> [[180]]#> lag(new_confirmed, n = 14) #> 0.5481366 #> #> [[181]]#> [1] NA#> #> [[182]]#> lag(new_confirmed, n = 14) #> -0.05410779 #> #> [[183]]#> lag(new_confirmed, n = 14) #> -0.007462687 #> #> [[184]]#> lag(new_confirmed, n = 14) #> 0.3680185 #> #> [[185]]#> lag(new_confirmed, n = 14) #> -0.00138019 #> #> [[186]]#> lag(new_confirmed, n = 14) #> 0.8943568 #> #> [[187]]#> lag(new_confirmed, n = 14) #> 0.4699798 #> #> [[188]]#> lag(new_confirmed, n = 14) #> -0.001714412 #> #> [[189]]#> lag(new_confirmed, n = 14) #> -0.01177239 #> #> [[190]]#> lag(new_confirmed, n = 14) #> 0.4459296 #> #> [[191]]#> lag(new_confirmed, n = 14) #> 0.741075 #> #> [[192]]#> lag(new_confirmed, n = 14) #> 0.7394529 #> #> [[193]]#> lag(new_confirmed, n = 14) #> 0.9754129 #> #> [[194]]#> lag(new_confirmed, n = 14) #> -0.004582951 #> #> [[195]]#> lag(new_confirmed, n = 14) #> 0.1134682 #> #> [[196]]#> lag(new_confirmed, n = 14) #> 0.8579614 #> #> [[197]]#> lag(new_confirmed, n = 14) #> 0.7022731 #> #> [[198]]#> lag(new_confirmed, n = 14) #> 0.0003349323 #> #> [[199]]#> lag(new_confirmed, n = 14) #> -0.01591102 #> #> [[200]]#> lag(new_confirmed, n = 14) #> 0.902218 #> #> [[201]]#> [1] NA#> #> [[202]]#> lag(new_confirmed, n = 14) #> 0.2367338 #> #> [[203]]#> lag(new_confirmed, n = 14) #> 0.6937724 #> #> [[204]]#> lag(new_confirmed, n = 14) #> 8.226172e-05 #> #> [[205]]#> lag(new_confirmed, n = 14) #> 0.7049068 #> #> [[206]]#> lag(new_confirmed, n = 14) #> 0.4773192 #> #> [[207]]#> [1] NA#> #> [[208]]#> [1] NA#> #> [[209]]#> lag(new_confirmed, n = 14) #> 0.2683079 #> #> [[210]]#> lag(new_confirmed, n = 14) #> 0.3773631 #> #> [[211]]#> lag(new_confirmed, n = 14) #> 0.09471127 #> #> [[212]]#> lag(new_confirmed, n = 14) #> 0.791494 #> #> [[213]]#> [1] NA#> #> [[214]]#> [1] NA#> #> [[215]]#> lag(new_confirmed, n = 14) #> 0.9127807 #> #> [[216]]#> lag(new_confirmed, n = 14) #> 0.6683978 #> #> [[217]]#> lag(new_confirmed, n = 14) #> 0.4777528 #> #> [[218]]#> lag(new_confirmed, n = 14) #> 0.289256 #> #> [[219]]#> lag(new_confirmed, n = 14) #> 0.09288966 #> #> [[220]]#> lag(new_confirmed, n = 14) #> 0.1472352 #> #> [[221]]#> [1] NA#> #> [[222]]#> lag(new_confirmed, n = 14) #> 0.8942314 #> #> [[223]]#> lag(new_confirmed, n = 14) #> 0.8124493 #> #> [[224]]#> lag(new_confirmed, n = 14) #> 0.001282686 #> #> [[225]]#> lag(new_confirmed, n = 14) #> 1.164049 #> #> [[226]]#> lag(new_confirmed, n = 14) #> -0.7243288 #> #> [[227]]#> lag(new_confirmed, n = 14) #> 0.9300546 #> #> [[228]]#> lag(new_confirmed, n = 14) #> NA #> #> [[229]]#> [1] NA#> #> [[230]]#> lag(new_confirmed, n = 14) #> 0.9314963 #> #> [[231]]#> [1] NA#> #> [[232]]#> lag(new_confirmed, n = 14) #> 0.3004533 #> #> [[233]]#> lag(new_confirmed, n = 14) #> 0.9587314 #> #> [[234]]#> lag(new_confirmed, n = 14) #> 0.2780126 #> #> [[235]]#> lag(new_confirmed, n = 14) #> 0.4839777 #> #> [[236]]#> lag(new_confirmed, n = 14) #> 0.4202595covid19_lstcol$data %>% map(safely_extract_slope) %>% map_dbl("result")#> [1] 6.920523e-01 6.435756e-01#> [3] 5.329582e-01 5.595175e-01#> [5] 5.196389e-01 7.375993e-01#> [7] 8.669078e-01 8.971555e-01#> [9] NA 5.299203e-01#> [11] 9.182422e-01 9.642226e-01#> [13] 2.903094e-01 NA#> [15] 8.760548e-01 8.306039e-01#> [17] 1.478383e-01 1.038319e+00#> [19] -1.417372e-01 3.760244e-01#> [21] 2.665434e-01 7.554900e-02#> [23] 6.108023e-01 6.446241e-01#> [25] 4.912720e-01 6.540097e-01#> [27] 4.138478e-01 5.732950e-01#> [29] 6.704973e-01 8.998348e-01#> [31] 5.736442e-02 5.809416e-01#> [33] 1.256359e-01 1.167159e-01#> [35] 9.418862e-01 NA#> [37] NA 2.580678e-01#> [39] 4.058524e-01 NA#> [41] 4.110214e-01 8.410514e-01#> [43] 6.855367e-01 1.374103e-01#> [45] 2.689623e-01 5.826228e-01#> [47] 1.004124e+00 6.459742e-01#> [49] 8.822818e-01 NA#> [51] 7.600437e-01 NA#> [53] -1.675946e-02 NA#> [55] 9.635528e-01 1.049798e+00#> [57] NA 5.009491e-01#> [59] 1.780097e-01 6.290575e-01#> [61] NA 5.348175e-02#> [63] 2.492007e-01 5.076202e-01#> [65] -1.289268e-02 1.628999e-01#> [67] 8.211096e-01 6.890107e-01#> [69] 4.747831e-01 NA#> [71] 1.370787e-01 8.498793e-01#> [73] 2.613858e-02 NA#> [75] 4.039257e-01 2.783185e-01#> [77] NA 8.992036e-01#> [79] 7.484860e-01 4.874482e-01#> [81] NA 9.351811e-01#> [83] NA 2.809673e-02#> [85] NA NA#> [87] 8.144507e-01 NA#> [89] 2.512326e-01 1.763243e-02#> [91] 5.299206e-01 4.224714e-01#> [93] -1.196319e-02 2.954188e-01#> [95] NA NA#> [97] 4.821869e-01 7.901768e-01#> [99] 8.979504e-01 9.487975e-01#> [101] 7.877892e-01 NA#> [103] 8.682548e-01 2.087615e-01#> [105] 8.144906e-01 7.282967e-01#> [107] 5.431636e-01 8.678327e-01#> [109] NA 1.177228e+00#> [111] 1.624997e-01 7.411408e-01#> [113] 2.179744e-02 9.123329e-01#> [115] 1.588501e-01 2.238890e-02#> [117] 9.392607e-01 7.137065e-01#> [119] 1.059572e-01 1.280151e-01#> [121] 7.095778e-01 8.902059e-01#> [123] 8.601565e-01 9.333330e-01#> [125] NA NA#> [127] 7.684239e-01 2.407153e-01#> [129] 8.741184e-01 8.370697e-01#> [131] 4.458898e-01 8.722886e-01#> [133] -1.612903e-02 NA#> [135] 7.781136e-01 4.789353e-01#> [137] NA 5.295625e-01#> [139] 8.784718e-01 4.523111e-01#> [141] 6.500141e-02 7.416119e-01#> [143] 8.947562e-01 7.780699e-01#> [145] -3.802281e-03 1.044826e+00#> [147] 9.334443e-01 5.462774e-03#> [149] 7.127593e-01 6.934141e-01#> [151] 4.480632e-01 7.526479e-01#> [153] 6.662816e-01 2.028236e-01#> [155] NA 9.219172e-01#> [157] NA 5.813632e-01#> [159] NA 7.849124e-01#> [161] 8.179260e-02 7.567328e-01#> [163] 4.486768e-01 9.711064e-01#> [165] 8.404479e-01 8.340365e-01#> [167] NA 1.146428e+00#> [169] NA NA#> [171] NA NA#> [173] NA 9.711078e-01#> [175] 9.645341e-01 NA#> [177] 1.992464e-01 NA#> [179] 5.725861e-02 5.481366e-01#> [181] NA -5.410779e-02#> [183] -7.462687e-03 3.680185e-01#> [185] -1.380190e-03 8.943568e-01#> [187] 4.699798e-01 -1.714412e-03#> [189] -1.177239e-02 4.459296e-01#> [191] 7.410750e-01 7.394529e-01#> [193] 9.754129e-01 -4.582951e-03#> [195] 1.134682e-01 8.579614e-01#> [197] 7.022731e-01 3.349323e-04#> [199] -1.591102e-02 9.022180e-01#> [201] NA 2.367338e-01#> [203] 6.937724e-01 8.226172e-05#> [205] 7.049068e-01 4.773192e-01#> [207] NA NA#> [209] 2.683079e-01 3.773631e-01#> [211] 9.471127e-02 7.914940e-01#> [213] NA NA#> [215] 9.127807e-01 6.683978e-01#> [217] 4.777528e-01 2.892560e-01#> [219] 9.288966e-02 1.472352e-01#> [221] NA 8.942314e-01#> [223] 8.124493e-01 1.282686e-03#> [225] 1.164049e+00 -7.243288e-01#> [227] 9.300546e-01 NA#> [229] NA 9.314963e-01#> [231] NA 3.004533e-01#> [233] 9.587314e-01 2.780126e-01#> [235] 4.839777e-01 4.202595e-01Alternatively, mutate() with map() inside a tibble
covid19_lstcol %>% mutate( slopes = map(data, safely_extract_slope) %>% map_dbl("result"))#> # A tibble: 236 x 3#> country_region data slopes#> <chr> <list> <dbl>#> 1 Afghanistan <tibble[,7] [306 × 7]> 0.692#> 2 Albania <tibble[,7] [298 × 7]> 0.644#> 3 Algeria <tibble[,7] [306 × 7]> 0.533#> 4 Andorra <tibble[,7] [305 × 7]> 0.560#> 5 Angola <tibble[,7] [287 × 7]> 0.520#> 6 Antigua and Barbuda <tibble[,7] [294 × 7]> 0.738#> # … with 230 more rowsSide-effects
Most functions are called for the value that they return. But some functions are called primarily for their side-effects, for example:
print()displays values/plots to the screen.write_csv()writes values to the CSV file saved in the disk.
No values/outputs to be captured.
We're going to save each region's data into separate CSV files to make it easier to import in the future.
group_split() splits tibble into chunks
covid19_lst <- covid19_byregion %>% group_by(country_region) %>% group_split()covid19_lst[[1]]#> # A tibble: 306 x 8#> country_region date confirmed deaths recovered#> <chr> <date> <dbl> <dbl> <dbl>#> 1 Afghanistan 2020-03-01 1 0 0#> 2 Afghanistan 2020-03-02 1 0 0#> 3 Afghanistan 2020-03-03 2 0 0#> 4 Afghanistan 2020-03-04 4 0 0#> 5 Afghanistan 2020-03-05 4 0 0#> 6 Afghanistan 2020-03-06 4 0 0#> # … with 300 more rows, and 3 more variables:#> # new_confirmed <dbl>, new_deaths <dbl>,#> # new_recovered <dbl>walk() with side-effects, if no outputs
region_names <- unique(covid19_byregion$country_region)file_name <- glue::glue("data/covid19/{region_names}.csv")# fs::dir_create("data/covid19")walk2(covid19_lst, file_name, write_csv)fs::dir_ls("data/covid19")#> data/covid19/Afghanistan.csv#> data/covid19/Albania.csv#> data/covid19/Algeria.csv#> data/covid19/Andorra.csv#> data/covid19/Angola.csv#> data/covid19/Antigua and Barbuda.csv#> data/covid19/Argentina.csv#> data/covid19/Armenia.csv#> data/covid19/Aruba.csv#> data/covid19/Australia.csv#> data/covid19/Austria.csv#> data/covid19/Azerbaijan.csv#> data/covid19/Bahamas, The.csv#> data/covid19/Bahamas.csv#> data/covid19/Bahrain.csv#> data/covid19/Bangladesh.csv#> data/covid19/Barbados.csv#> data/covid19/Belarus.csv#> data/covid19/Belgium.csv#> data/covid19/Belize.csv#> data/covid19/Benin.csv#> data/covid19/Bhutan.csv#> data/covid19/Bolivia.csv#> data/covid19/Bosnia and Herzegovina.csv#> data/covid19/Botswana.csv#> data/covid19/Brazil.csv#> data/covid19/Brunei.csv#> data/covid19/Bulgaria.csv#> data/covid19/Burkina Faso.csv#> data/covid19/Burma.csv#> data/covid19/Burundi.csv#> data/covid19/Cabo Verde.csv#> data/covid19/Cambodia.csv#> data/covid19/Cameroon.csv#> data/covid19/Canada.csv#> data/covid19/Cape Verde.csv#> data/covid19/Cayman Islands.csv#> data/covid19/Central African Republic.csv#> data/covid19/Chad.csv#> data/covid19/Channel Islands.csv#> data/covid19/Chile.csv#> data/covid19/China.csv#> data/covid19/Colombia.csv#> data/covid19/Comoros.csv#> data/covid19/Congo (Brazzaville).csv#> data/covid19/Congo (Kinshasa).csv#> data/covid19/Costa Rica.csv#> data/covid19/Cote d'Ivoire.csv#> data/covid19/Croatia.csv#> data/covid19/Cruise Ship.csv#> data/covid19/Cuba.csv#> data/covid19/Curacao.csv#> data/covid19/Cyprus.csv#> data/covid19/Czech Republic.csv#> data/covid19/Czechia.csv#> data/covid19/Denmark.csv#> data/covid19/Diamond Princess.csv#> data/covid19/Djibouti.csv#> data/covid19/Dominica.csv#> data/covid19/Dominican Republic.csv#> data/covid19/East Timor.csv#> data/covid19/Ecuador.csv#> data/covid19/Egypt.csv#> data/covid19/El Salvador.csv#> data/covid19/Equatorial Guinea.csv#> data/covid19/Eritrea.csv#> data/covid19/Estonia.csv#> data/covid19/Eswatini.csv#> data/covid19/Ethiopia.csv#> data/covid19/Faroe Islands.csv#> data/covid19/Fiji.csv#> data/covid19/Finland.csv#> data/covid19/France.csv#> data/covid19/French Guiana.csv#> data/covid19/Gabon.csv#> data/covid19/Gambia, The.csv#> data/covid19/Gambia.csv#> data/covid19/Georgia.csv#> data/covid19/Germany.csv#> data/covid19/Ghana.csv#> data/covid19/Gibraltar.csv#> data/covid19/Greece.csv#> data/covid19/Greenland.csv#> data/covid19/Grenada.csv#> data/covid19/Guadeloupe.csv#> data/covid19/Guam.csv#> data/covid19/Guatemala.csv#> data/covid19/Guernsey.csv#> data/covid19/Guinea-Bissau.csv#> data/covid19/Guinea.csv#> data/covid19/Guyana.csv#> data/covid19/Haiti.csv#> data/covid19/Holy See.csv#> data/covid19/Honduras.csv#> data/covid19/Hong Kong SAR.csv#> data/covid19/Hong Kong.csv#> data/covid19/Hungary.csv#> data/covid19/Iceland.csv#> data/covid19/India.csv#> data/covid19/Indonesia.csv#> data/covid19/Iran (Islamic Republic of).csv#> data/covid19/Iran.csv#> data/covid19/Iraq.csv#> data/covid19/Ireland.csv#> data/covid19/Israel.csv#> data/covid19/Italy.csv#> data/covid19/Jamaica.csv#> data/covid19/Japan.csv#> data/covid19/Jersey.csv#> data/covid19/Jordan.csv#> data/covid19/Kazakhstan.csv#> data/covid19/Kenya.csv#> data/covid19/Kosovo.csv#> data/covid19/Kuwait.csv#> data/covid19/Kyrgyzstan.csv#> data/covid19/Laos.csv#> data/covid19/Latvia.csv#> data/covid19/Lebanon.csv#> data/covid19/Lesotho.csv#> data/covid19/Liberia.csv#> data/covid19/Libya.csv#> data/covid19/Liechtenstein.csv#> data/covid19/Lithuania.csv#> data/covid19/Luxembourg.csv#> data/covid19/MS Zaandam.csv#> data/covid19/Macao SAR.csv#> data/covid19/Macau.csv#> data/covid19/Madagascar.csv#> data/covid19/Malawi.csv#> data/covid19/Malaysia.csv#> data/covid19/Maldives.csv#> data/covid19/Mali.csv#> data/covid19/Malta.csv#> data/covid19/Marshall Islands.csv#> data/covid19/Martinique.csv#> data/covid19/Mauritania.csv#> data/covid19/Mauritius.csv#> data/covid19/Mayotte.csv#> data/covid19/Mexico.csv#> data/covid19/Moldova.csv#> data/covid19/Monaco.csv#> data/covid19/Mongolia.csv#> data/covid19/Montenegro.csv#> data/covid19/Morocco.csv#> data/covid19/Mozambique.csv#> data/covid19/Namibia.csv#> data/covid19/Nepal.csv#> data/covid19/Netherlands.csv#> data/covid19/New Zealand.csv#> data/covid19/Nicaragua.csv#> data/covid19/Niger.csv#> data/covid19/Nigeria.csv#> data/covid19/North Macedonia.csv#> data/covid19/Norway.csv#> data/covid19/Oman.csv#> data/covid19/Others.csv#> data/covid19/Pakistan.csv#> data/covid19/Palestine.csv#> data/covid19/Panama.csv#> data/covid19/Papua New Guinea.csv#> data/covid19/Paraguay.csv#> data/covid19/Peru.csv#> data/covid19/Philippines.csv#> data/covid19/Poland.csv#> data/covid19/Portugal.csv#> data/covid19/Puerto Rico.csv#> data/covid19/Qatar.csv#> data/covid19/Republic of Ireland.csv#> data/covid19/Republic of Korea.csv#> data/covid19/Republic of Moldova.csv#> data/covid19/Republic of the Congo.csv#> data/covid19/Reunion.csv#> data/covid19/Romania.csv#> data/covid19/Russia.csv#> data/covid19/Russian Federation.csv#> data/covid19/Rwanda.csv#> data/covid19/Saint Barthelemy.csv#> data/covid19/Saint Kitts and Nevis.csv#> data/covid19/Saint Lucia.csv#> data/covid19/Saint Martin.csv#> data/covid19/Saint Vincent and the Grenadines.csv#> data/covid19/Samoa.csv#> data/covid19/San Marino.csv#> data/covid19/Sao Tome and Principe.csv#> data/covid19/Saudi Arabia.csv#> data/covid19/Senegal.csv#> data/covid19/Serbia.csv#> data/covid19/Seychelles.csv#> data/covid19/Sierra Leone.csv#> data/covid19/Singapore.csv#> data/covid19/Slovakia.csv#> data/covid19/Slovenia.csv#> data/covid19/Solomon Islands.csv#> data/covid19/Somalia.csv#> data/covid19/South Africa.csv#> data/covid19/South Korea.csv#> data/covid19/South Sudan.csv#> data/covid19/Spain.csv#> data/covid19/Sri Lanka.csv#> data/covid19/St. Martin.csv#> data/covid19/Sudan.csv#> data/covid19/Suriname.csv#> data/covid19/Sweden.csv#> data/covid19/Switzerland.csv#> data/covid19/Syria.csv#> data/covid19/Taipei and environs.csv#> data/covid19/Taiwan*.csv#> data/covid19/Taiwan.csv#> data/covid19/Tajikistan.csv#> data/covid19/Tanzania.csv#> data/covid19/Thailand.csv#> data/covid19/The Bahamas.csv#> data/covid19/The Gambia.csv#> data/covid19/Timor-Leste.csv#> data/covid19/Togo.csv#> data/covid19/Trinidad and Tobago.csv#> data/covid19/Tunisia.csv#> data/covid19/Turkey.csv#> data/covid19/UK.csv#> data/covid19/US.csv#> data/covid19/Uganda.csv#> data/covid19/Ukraine.csv#> data/covid19/United Arab Emirates.csv#> data/covid19/United Kingdom.csv#> data/covid19/Uruguay.csv#> data/covid19/Uzbekistan.csv#> data/covid19/Vanuatu.csv#> data/covid19/Vatican City.csv#> data/covid19/Venezuela.csv#> data/covid19/Viet Nam.csv#> data/covid19/Vietnam.csv#> data/covid19/West Bank and Gaza.csv#> data/covid19/Yemen.csv#> data/covid19/Zambia.csv#> data/covid19/Zimbabwe.csv#> data/covid19/occupied Palestinian territory.csv{fs} provides a cross-platform, uniform interface to file system operations.
library(fs)covid19_local <- map_dfr( dir_ls("data/covid19", glob = "*.csv"), read_csv)covid19_local#> # A tibble: 56,107 x 8#> country_region date confirmed deaths recovered#> <chr> <date> <dbl> <dbl> <dbl>#> 1 Afghanistan 2020-03-01 1 0 0#> 2 Afghanistan 2020-03-02 1 0 0#> 3 Afghanistan 2020-03-03 2 0 0#> 4 Afghanistan 2020-03-04 4 0 0#> 5 Afghanistan 2020-03-05 4 0 0#> 6 Afghanistan 2020-03-06 4 0 0#> # … with 56,101 more rows, and 3 more variables:#> # new_confirmed <dbl>, new_deaths <dbl>,#> # new_recovered <dbl>