Lab 07 Solution
This lab exercise is due 23:59 Monday 10 May (NZST).
- You should submit an R file (i.e. file extension
.R) containing R code that assigns the appropriate values to the appropriate symbols. - Your R file will be executed in order and checked against the values that have been assigned to the symbols using an automatic grading system. Marks will be fully deducted for non-identical results.
- Intermediate steps to achieve the final results will NOT be checked.
- Each question is worth 0.2 points.
- You should submit your R file on Canvas.
- Late assignments are NOT accepted unless prior arrangement for medical/compassionate reasons.
In this lab exercise, you are going to work on the alternative
visualisation of “Flattening the
curve” for the publication purpose,
using covid19-daily-cases.csv that contains cumulative daily confirmed
cases between 2020-03-01 and 2020-05-31 around the world. You shall use
the following code snippet (and include them upfront in your R file)
for this lab session:
library(scales)
library(tidyverse)
selected <- c("Australia", "Brazil", "Canada", "China", "France", "Germany",
"India", "Israel", "Italy", "Japan", "Korea, South", "New Zealand",
"Spain", "Sweden", "United Kingdom", "US")
covid19 <- read_csv("data/covid19-daily-cases.csv") %>%
filter(country_region %in% selected)
covid19
#> # A tibble: 1,452 x 3
#> country_region date confirmed
#> <chr> <date> <dbl>
#> 1 Australia 2020-03-01 27
#> 2 Australia 2020-03-02 30
#> 3 Australia 2020-03-03 39
#> 4 Australia 2020-03-04 52
#> 5 Australia 2020-03-05 55
#> 6 Australia 2020-03-06 60
#> 7 Australia 2020-03-07 63
#> 8 Australia 2020-03-08 76
#> 9 Australia 2020-03-09 91
#> 10 Australia 2020-03-10 107
#> # … with 1,442 more rows
Suppose that you have created an Rproj for this course. You need to
download covid19-daily-cases.csv
here,
to data/ under your Rproj folder.
- You’re required to use relative file paths
data/covid19-daily-cases.csvto import these data. NO marks will be given to this lab for using URL links or different file paths.
Question 1
Substitute "Korea, South" with "South Korea" in country_region,
and then add a new column new_cases that computes new confirmed cases
in the past week for each region.
You should end up with a tibble called covid19_cases.
HINTS
- You may find one of {dplyr}’s vector functions useful.
covid19_cases <- covid19 %>%
mutate(country_region = case_when(
country_region == "Korea, South" ~ "South Korea",
TRUE ~ country_region)) %>%
group_by(country_region) %>%
mutate(new_cases = confirmed - lag(confirmed, 7)) %>%
ungroup()
covid19_cases
#> # A tibble: 1,452 x 4
#> country_region date confirmed new_cases
#> <chr> <date> <dbl> <dbl>
#> 1 Australia 2020-03-01 27 NA
#> 2 Australia 2020-03-02 30 NA
#> 3 Australia 2020-03-03 39 NA
#> 4 Australia 2020-03-04 52 NA
#> 5 Australia 2020-03-05 55 NA
#> 6 Australia 2020-03-06 60 NA
#> 7 Australia 2020-03-07 63 NA
#> 8 Australia 2020-03-08 76 49
#> 9 Australia 2020-03-09 91 61
#> 10 Australia 2020-03-10 107 68
#> # … with 1,442 more rows
mean(covid19_cases$new_cases, na.rm = TRUE)
#> [1] 18871.79
Question 2
Rename country_region to country_region_bg.
You should end up with a tibble called covid19_bg.
covid19_bg <- covid19_cases %>%
rename(country_region_bg = country_region)
covid19_bg
#> # A tibble: 1,452 x 4
#> country_region_bg date confirmed new_cases
#> <chr> <date> <dbl> <dbl>
#> 1 Australia 2020-03-01 27 NA
#> 2 Australia 2020-03-02 30 NA
#> 3 Australia 2020-03-03 39 NA
#> 4 Australia 2020-03-04 52 NA
#> 5 Australia 2020-03-05 55 NA
#> 6 Australia 2020-03-06 60 NA
#> 7 Australia 2020-03-07 63 NA
#> 8 Australia 2020-03-08 76 49
#> 9 Australia 2020-03-09 91 61
#> 10 Australia 2020-03-10 107 68
#> # … with 1,442 more rows
Question 3
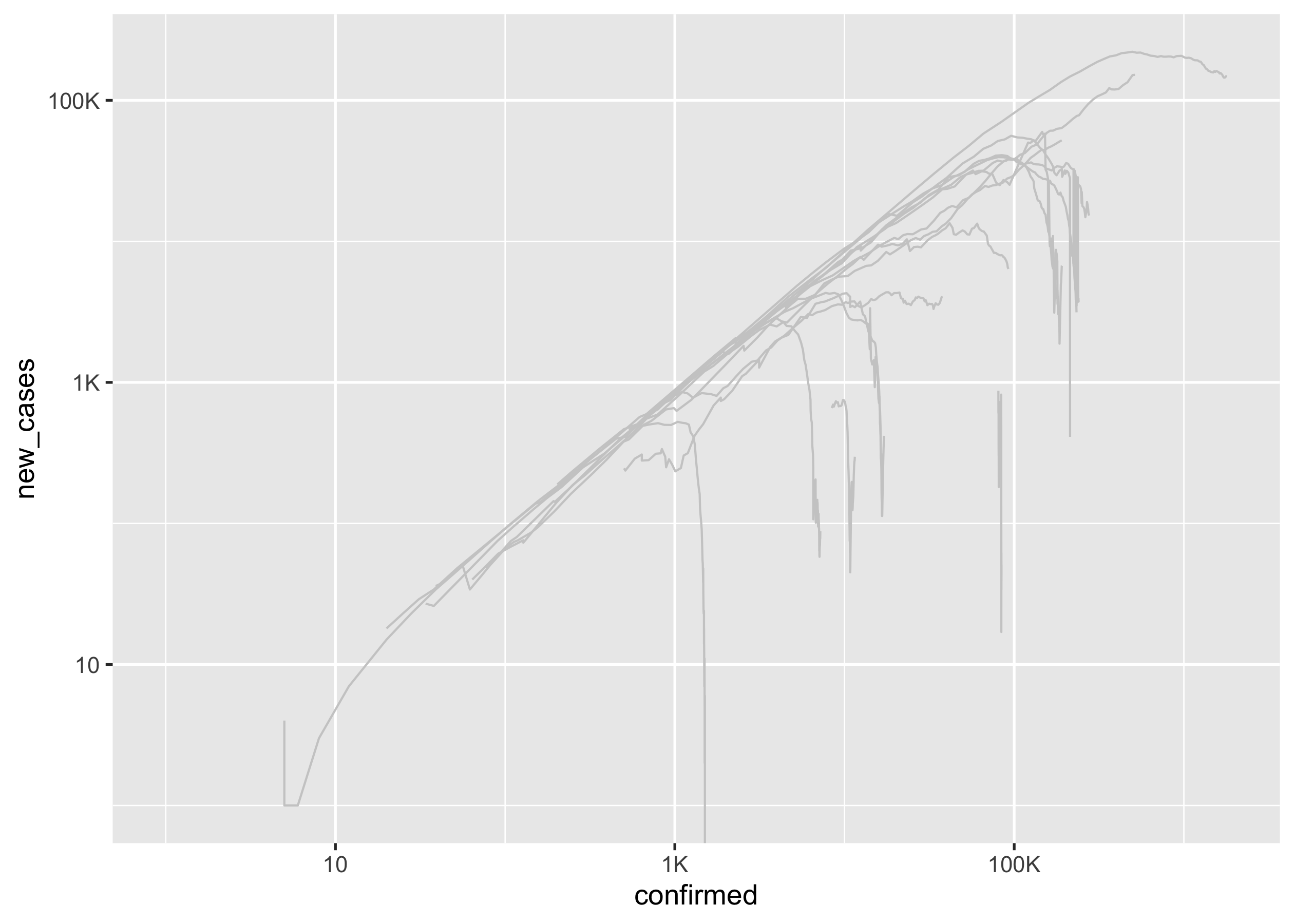
Create a line chart of new_cases vs confirmed for each region.
You should end up with a ggplot called p1, with
- lines:
colour = "gray80"andsize = 0.4, label_number_si()to axis labels and logarithmic scales applied.
p1 <- covid19_bg %>%
ggplot(aes(x = confirmed, y = new_cases)) +
geom_line(aes(group = country_region_bg), colour = "gray80", size = 0.4) +
scale_x_log10(labels = label_number_si()) +
scale_y_log10(labels = label_number_si())
p1
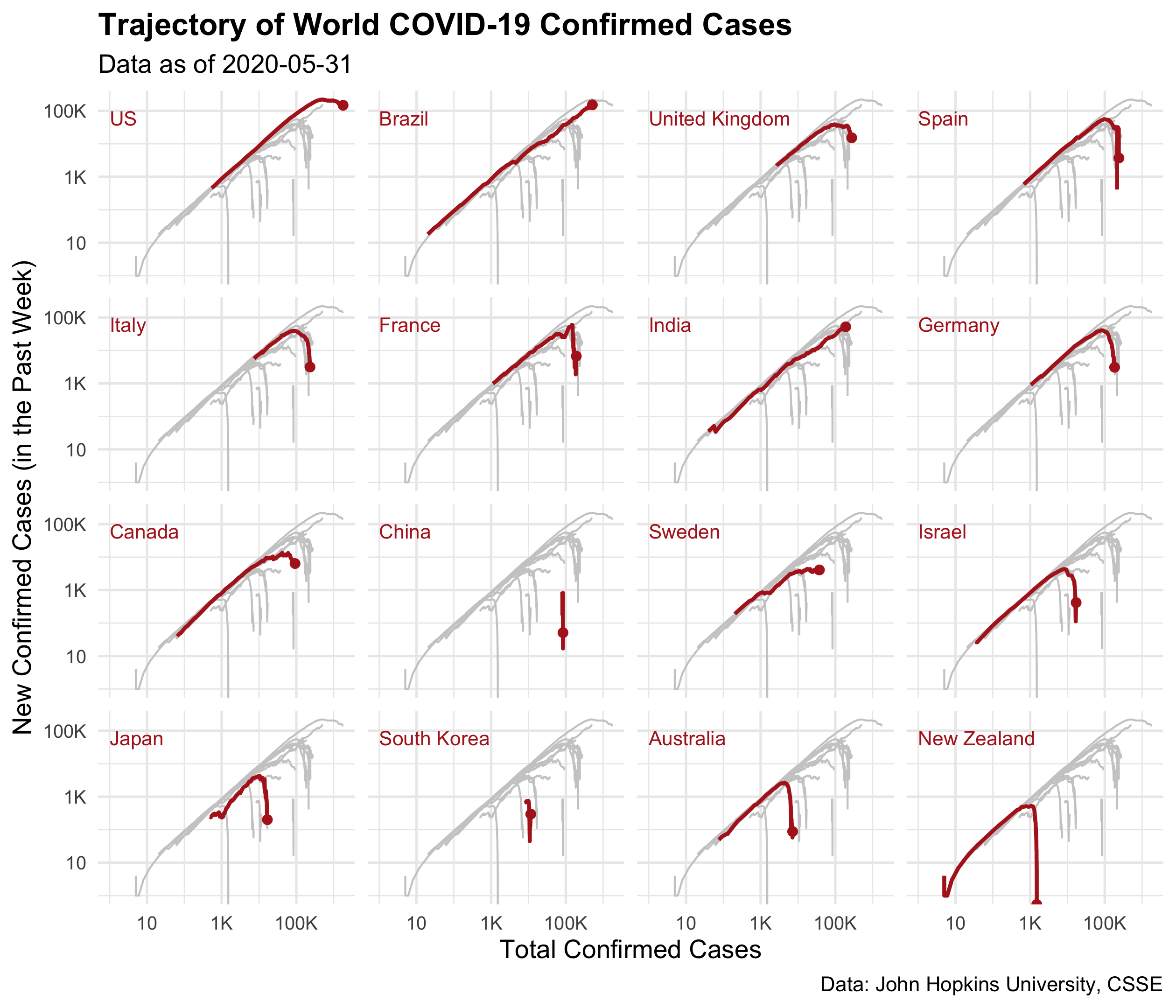
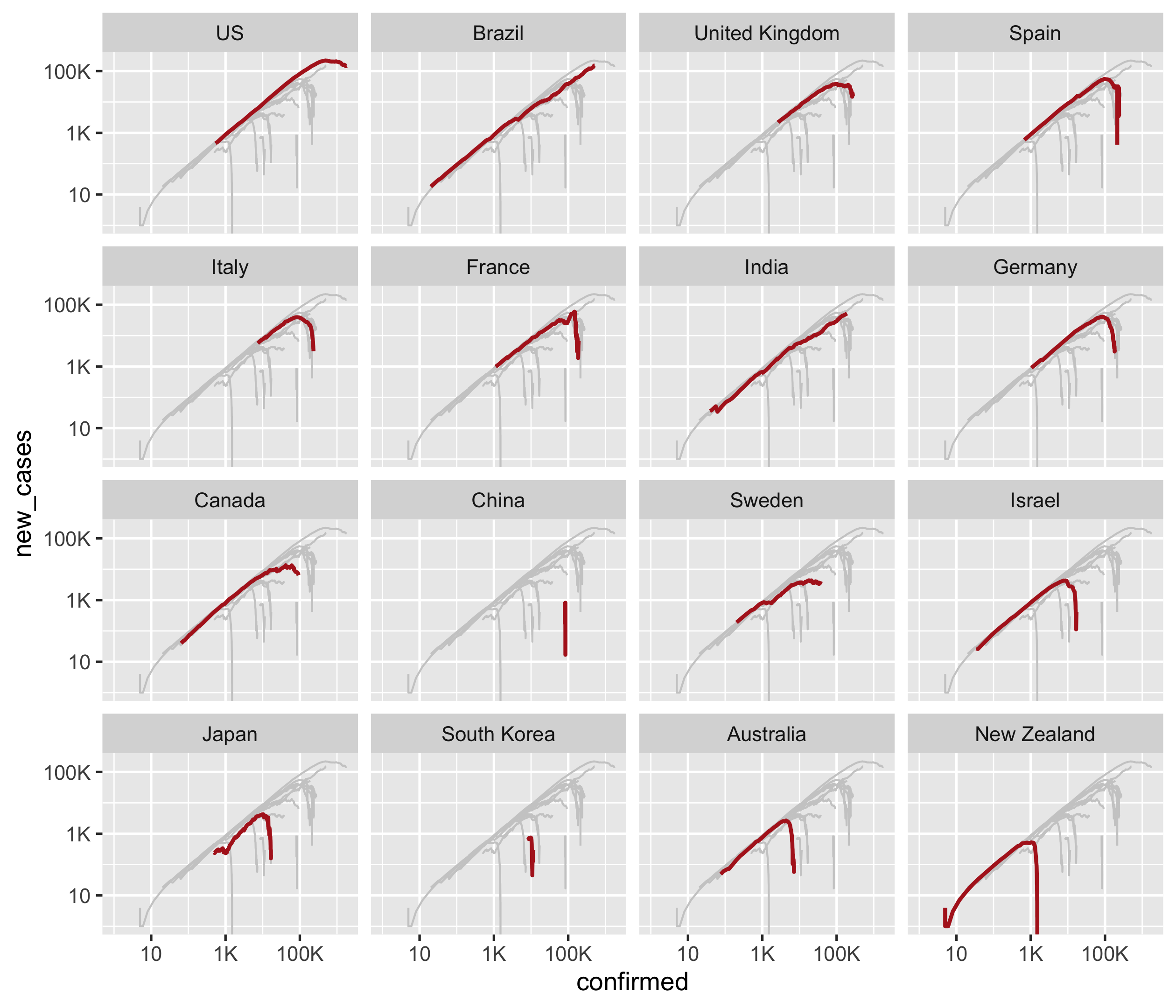
Question 4
Generate a small multiples of p1 by regions ordered by their confirmed
cases on 2020-05-31 from high to low, and highlight each region with
colour = "firebrick" and size = 0.8 for each panel.
You should end up with a ggplot called p2.
p2 <- p1 +
geom_line(colour = "firebrick", size = 0.8, data = covid19_cases) +
facet_wrap(~ fct_reorder(country_region, -confirmed, .fun = last))
p2
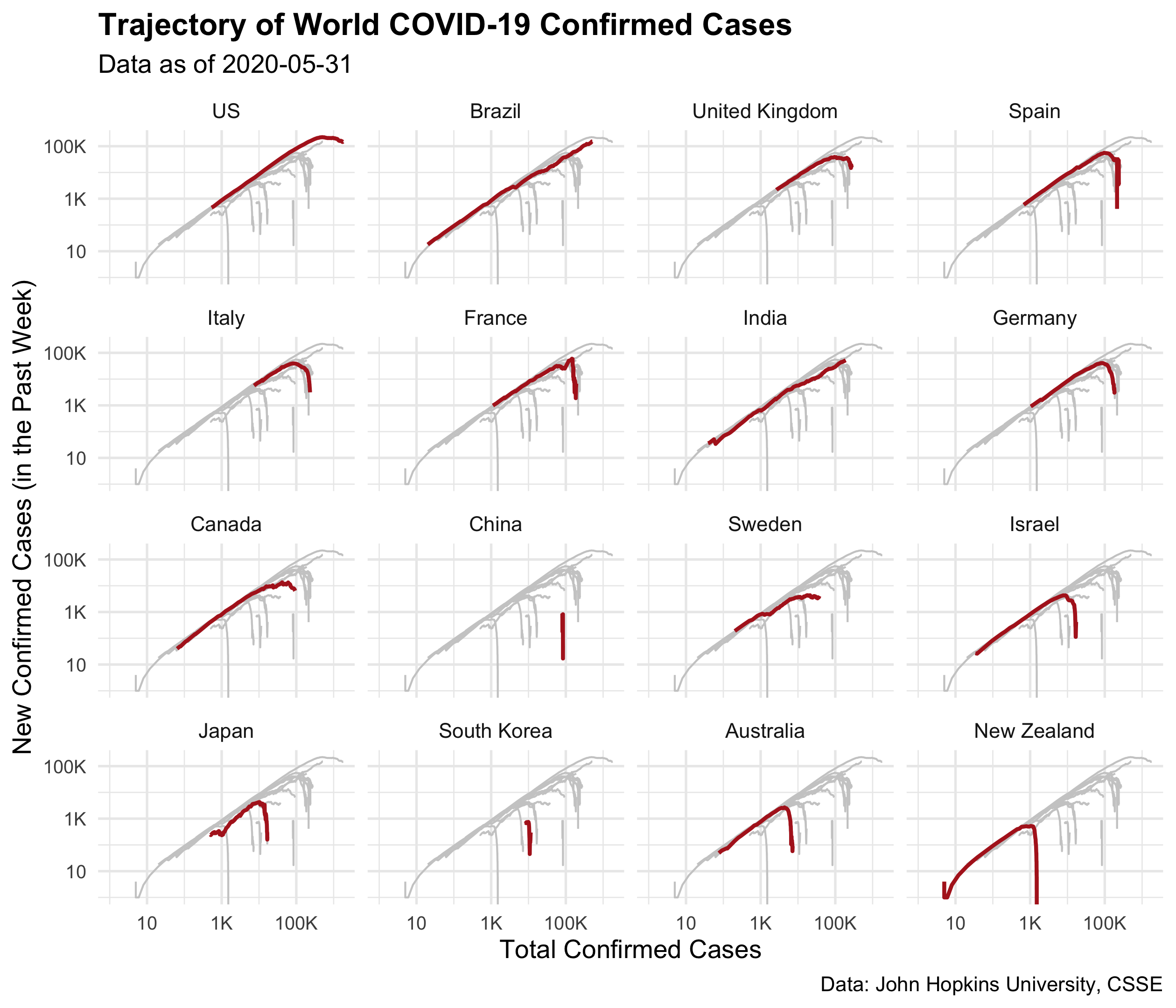
Question 5
Add the following titles and labels to p2:
- x:
"Total Confirmed Cases", - y:
"New Confirmed Cases (in the Past Week)", - title:
"Trajectory of World COVID-19 Confirmed Cases", - subtitle:
"Data as of 2020-05-31", - caption:
"Data: John Hopkins University, CSSE"
You should end up with a ggplot called p3, with
- the minimal theme,
- the plot title in bold,
- setting the axis text font size to be 8.
p3 <- p2 +
labs(
x = "Total Confirmed Cases",
y = "New Confirmed Cases (in the Past Week)",
title = "Trajectory of World COVID-19 Confirmed Cases",
subtitle = "Data as of 2020-05-31",
caption = "Data: John Hopkins University, CSSE") +
theme_minimal() +
theme(
plot.title = element_text(face = "bold"),
axis.text = element_text(size = 8)
)
p3
Question4fun (NO marks)
Polish p3 by:
- adding the point at the end of each highlighted line,
- placing each region labels at the top left position of each panel,
- removing facet texts.
covid19_end <- covid19_cases %>%
group_by(country_region) %>%
summarise(
confirmed = max(confirmed),
new_cases = last(new_cases)
)
covid19_text <- covid19_cases %>%
group_by(country_region) %>%
summarise(confirmed = 1, new_cases = 100000)
p3 +
geom_point(data = covid19_end, colour = "firebrick") +
geom_text(aes(label = country_region), data = covid19_text,
vjust = "inward", hjust = "inward", colour = "firebrick", size = 3) +
theme(strip.text = element_blank())